the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
BGC-val: a model- and grid-independent Python toolkit to evaluate marine biogeochemical models
Andrew Yool
Julien Palmieri
Alistair Sellar
Till Kuhlbrodt
Ekaterina Popova
Colin Jones
J. Icarus Allen
The biogeochemical evaluation toolkit, BGC-val, is a model- and grid-independent Python toolkit that has been built to evaluate marine biogeochemical models using a simple interface. Here, we present the ideas that motivated the development of the BGC-val software framework, introduce the code structure, and show some applications of the toolkit using model results from the Fifth Climate Model Intercomparison Project (CMIP5). A brief outline of how to access and install the repository is presented in Appendix A, but the specific details on how to use the toolkit are kept in the code repository.
The key ideas that directed the toolkit design were model and grid independence, front-loading analysis functions and regional masking, interruptibility, and ease of use. We present each of these goals, why they were important, and what we did to address them. We also present an outline of the code structure of the toolkit illustrated with example plots produced by the toolkit.
After describing BGC-val, we use the toolkit to investigate the performance of the marine physical and biogeochemical quantities of the CMIP5 models and highlight some predictions about the future state of the marine ecosystem under a business-as-usual CO2 concentration scenario (RCP8.5).
- Article
(3325 KB) - Full-text XML
-
Supplement
(14874 KB) - BibTeX
- EndNote
It is widely known that climate change is expected to have a significant impact on weather patterns, the cryosphere, the land surface, and the ocean (Cook et al., 2013; Le Quéré et al., 2013; Rhein et al., 2013; Stocker et al., 2015). Marine organisms are vulnerable not only to impacts of rising temperatures, but also to the associated deoxygenation (Gruber, 2011; Stramma et al., 2008) and ocean acidification driven by ocean CO2 uptake (Azevedo et al., 2015; Caldeira and Wickett, 2003; Dutkiewicz et al., 2015). The ocean is an important sink of carbon, absorbing approximately 27 % of the anthropogenic carbon emitted between 2002 and 2011 (Le Quéré et al., 2013). Under a changing climate, the ocean is likely to continue to absorb some of the anthropogenic atmospheric carbon dioxide, rendering the ocean more acidic via the increased formation of carbonic acid. The acidification of the ocean is expected to continue to have a significant impact on sea life (Dutkiewicz et al., 2015; Rhein et al., 2013). Due to the high thermal capacity of water, nearly all of the excess heat captured by the greenhouse effect is absorbed by the ocean (Rhein et al., 2013). This increases the temperature of the waters, which causes sea levels to rise via thermal expansion (Church et al., 2013), may accelerate the melting of sea ice (Moore et al., 2015), and may push many marine organisms outside of their thermal tolerance range (Poloczanska et al., 2016).
The 2016 Paris Climate Accord is a wide-ranging international agreement on greenhouse gas emissions mitigation and climate change adaptation which is underpinned by the goal of limiting the global mean temperature increase to less than 2 ∘C above pre-industrial levels (Schleussner et al., 2016). International environmental policies like the Paris Climate Accord hinge on the projections made by the scientific community. Numerical models of the Earth system are the only tools available to make meaningful predictions about the future of our climate. However, in order to trust the results of the models, they must first be demonstrated to be a sufficiently good representation of the Earth system. The process of testing the behaviour of the simulations is known as model evaluation. The importance of evaluating the models grows in significance as models are increasingly used to inform policy (Brown and Caldeira, 2017).
The Coupled Model Intercomparison Project (CMIP) is a framework for coordinating climate change experiments and providing information of value to the International Panel on Climate Change (IPCC) Working Groups (Taylor et al., 2012). CMIP5 was set up to address outstanding scientific questions, to improve understanding of climate, and to provide estimates of future climate change that will be useful to those considering its possible consequences (Meehl et al., 2009; Taylor et al., 2007). These models represent the best scientific projections of the range of possible climates going into the 21st century. The results of previous rounds of CMIP comparisons have become a crucial component of the IPCC reports. In the fifth phase of CMIP, many of the climate forecasts were based on representative concentration pathways (RCPs), which represented different possibilities for greenhouse gas concentrations in the 21st century (Moss et al., 2010; van Vuuren et al., 2011).
The upcoming Sixth Climate Model Intercomparison Project, CMIP6 (Eyring et al., 2016), is expected to start receiving models in the year 2018. In order to contribute to CMIP6, each model must complete a suite of scenarios known as the Diagnosis, Evaluation, and Characterisation of Klima (DECK) simulations, which include an atmospheric model intercomparison between the years 1979 and 2014, a pre-industrial control simulation, a 1 % per year CO2 increase, an abrupt 4×CO2 run, and a historical simulation using CMIP6 forcings (1850–2014). CMIP6 models are also required to use consistent standardisation, coordination, infrastructure, and documentation.
Numerical simulations are the only tools available to predict how rising temperature, atmospheric CO2, and other factors will influence marine life in the future. Furthermore, Earth system models are also the tool that can project how changes in the marine system feed back on and interact with other climate-relevant components of the Earth system. The UK Earth system model (UKESM1) is a next-generation Earth system model currently under development. The aim of UKESM1 is to develop and apply a world-leading Earth system model. Simulations made with the UKESM1 will be contributed to CMIP6.
During the process of building the UKESM1, we also deployed a suite of tools to monitor the marine component of the model as it was being developed. This software suite is called BGC-val, and it is used to compare the marine components of simulations against each other and against observational data, with an emphasis on marine biogeochemistry. The suite of evaluation tools that we present in this work is a generalised extension of those tools. BGC-val has been deployed operationally since June 2016 and has been used extensively for the development, evaluation, and tuning of the spin-up and CMIP6 DECK runs of the marine component of the UKESM1, MEDUSA (Yool et al., 2013). The earliest version of BGC-val was based on the tools used to evaluate the development of the NEMO-ERSEM simulations in the iMarNet project (Kwiatkowski et al., 2014).
The focus of this work is not to prepare a guide on how to run the BGC-val
code, but rather to present the central ideas and methods used to design the
toolkit. A brief description of how to install, set up, and run the code can
be found in the Appendix A. Further details are available
in the README.md file, which can be found in the base directory of
the code repository. Alternatively, the README.md file can be viewed
by registered users on the landing page of the BGC-val toolkit GitLab server.
Instructions on how to register and access the toolkit can be found below in
the “Code availability” section. After this introduction,
Sect. 2 outlines the features of the BGC-val toolkit,
Sect. 3 describes the evaluation process used by the toolkit,
and Sect. 4 describes the code structure of the BGC-val
toolkit. Finally, Sect. 5 shows some examples of the toolkit
in use with model data from CMIP5.
Model evaluation tools
The evaluation of marine ecosystem models is a crucial stage in the deployment of climate models to inform policy decisions. When compared to models of other parts of the Earth system, marine models have several unique features which complicate the model evaluation process. The data available for evaluating a marine model can be relatively scarce. The ocean covers more than twice as much of the surface of the Earth than land, and there are sizeable regions of the ocean which are rarely visited by humans, let alone sampled by scientific cruises (Garcia-Castellanos and Lombardo, 2007). In addition, only the surface of the ocean is visible to satellites; the properties of marine life in the deep waters cannot be observed from remote sensing. Similarly, the connections between different components of the Earth system can also be difficult to measure. Several crucial global fluxes are unknown or estimated with significant uncertainties, such as the global total flux of CO2 into the ocean (Takahashi et al., 2009), the global total deposition of atmospheric dust (Mahowald et al., 2005), or the global production export flux (Boyd and Trull, 2007; Henson et al., 2011). Prior to the development of BGC-val, there was no evaluation toolkit specific to models of the marine ecosystem and evaluation was typically performed in an ad hoc manner.
As part of the preparation for CMIP6, a community diagnostic toolkit for evaluating climate models, ESMValTool, has been developed (Poloczanska et al., 2016). Like BGC-val, ESMValTool is a flexible Python-based model evaluation toolkit and many of the features developed for BGC-val also appear in ESMValTool. However, BGC-val was developed explicitly for evaluating models of the ocean, whereas ESMValTool was built to evaluate models of the entire Earth system. It must be noted that ESMValTool was not yet available for operational deployment when we started evaluating the UKESM1 spin in June 2016. Furthermore, ESMValTool contained very few ocean and marine biogeochemistry performance metrics at that point. The authors of ESMValTool are currently in the process of preparing ESMValTool version 2 for release in the autumn of 2018. This is a rapidly developing package, with several authors adding new features every week, and it is not likely to be finalised for operational deployment for several more months. However, many of the features that were implemented in BGC-val have since also been added to ESMValTool and the authors of BGC-val are also contributors to ESMValTool. In addition, many of the metrics deployed in BGC-val's ocean-specific evaluation have been proposed as key metrics to include in future versions of ESMValTool. A full description and access to the ESMValTool code is available via the GitHub page: https://github.com/ESMValGroup/ESMValTool (last access: 5 October 2018).
Marine Assess (formerly Ocean Assess) is a UK Met Office software toolkit for evaluating the physical circulation of the models developed there. From the authors' hands-on experience with Marine Assess, several of its metrics were specific to the NEMO ORCA1 grid, so it could not be deployed to evaluate the other CMIP5 models. Furthermore, Marine Assess is not available outside the UK Met Office and is not yet described in any public documentation. For these reasons, while Marine Assess is a powerful tool, it has yet to be embraced by the wider model evaluation community (Daley Calvert and Tim Graham, Marine Assess authors, Met Office UK, personal communication, 2018).
Outside of the marine environment, several toolkits are also available for evaluating models of the other parts of the Earth system. For the land surface, the Land surface Verification Toolkit (LVT) and the International Land Model Benchmarking (ILAMB) frameworks are available (Hoffman et al., 2017; Kumar et al., 2012). For the atmosphere, several packages are available, for instance the Atmospheric Model Evaluation Tool (AMET) (Appel et al., 2011), the Chemistry–Climate Model Validation Diagnostic (CCMVal-Diag) tool (Gettelman et al., 2012), or the Model Evaluation Tools (MET) (Fowler et al., 2018). Please note that these are not complete lists of the tools available.
Also note that in this work, we do not aim to introduce any new metrics or statistical methods. There are already plenty of valuable metrics and method descriptions available (Jolliff et al., 2009; Saux Picart et al., 2012; Stow et al., 2009; Taylor, 2001; de Mora et al., 2013, 2016).
In addition to the statistical tools available, the marine biogeochemistry community has access to many observational datasets. BGC-val has been used to compare various ocean models against a wide range of marine datasets, including the Takahashi air–sea flux of CO2 (Takahashi et al., 2009), the European Space Agency Climate Change Initiative (ESA-CCI) ocean colour dataset (Grant et al., 2017), the World Ocean Atlas data for temperature (Locarnini et al., 2013), salinity (Zweng et al., 2013), oxygen (Garcia et al., 2013a), and nutrients (Garcia et al., 2013b), and the MAREDAT (Buitenhuis et al., 2013b) global database for marine pigment (Peloquin et al., 2013), picophytoplankton (Buitenhuis et al., 2012), diatoms (Leblanc et al., 2012), and mesozooplankton (Moriarty and O'Brien, 2013). These datasets are all publicly available and are typically distributed as a monthly climatology or annual mean NetCDF file.
While BGC-val was originally built as a toolkit for investigating the time development of the marine biogeochemistry component of the UK Earth system model, UKESM1, the primary focus of BGC-val's development was to make the toolkits as generic as possible. This means that the tools can be easily adapted for use with for a wide range of models, spatial domains, model grids, fields, datasets, and timescales without needing significant changes to the underlying software and without any significant post-processing of the model or observational data. The toolkit was built to be model independent, grid independent, interruptible, simple to use, and to include front-loading analyses and masking functionality.
The BGC-val toolkit was written in Python 2.7. The reason that Python was used is because it is freely available and widely distributed; it is portable and available with most operating systems, and there are many powerful standard packages that can be easily imported or installed locally. It is object oriented (allowing front-loading functionality described below), and it is popular and hence well documented and well supported.
2.1 Model independence
The Earth system models submitted to CMIP5 were created by largely
independent groups of scientists. While some model developers build CMIP
compliance into their models, other model developers choose to use their
in-house style, then reformat the file names and contents to a uniform naming
and units scheme before they are submitted to CMIP. This flexibility means
that each model working group may use their own file-naming conventions,
dimension names, variables, and variable names until the data are submitted to
CMIP5. Outside of the CMIP5 standardisation, there are many competing
nomenclatures. For instance, in addition to the CMIP standard name,
lat, we have encountered the following nonstandard names in model
data files, all describing the latitude coordinate: lats,
rlat, nav_lat, latitude, and several other
variants. Similarly, different models and observational datasets may not
necessarily use the same units.
While the CMIP5 data have been produced using a uniform naming scheme, this toolkit allows for models to be evaluated without any prior assumptions on their naming conventions or units. This means that it would be possible to deploy this toolkit during the development stage of a model, before reformatting the data to CMIP compliance. This is how this toolkit was applied during the development of UKESM1. Model independency ensures that the toolkit can be applied in a range of scenarios, without requiring significant knowledge of the toolkit's inner workings and without post-processing the data.
2.2 Grid independence
For each Earth system model submitted to CMIP5, the development team chose how they wanted to divide the ocean into a grid composed of individual cells. Furthermore, unlike the naming and unit schemes, the model data submitted to CMIP5 have not been reformatted to a uniform grid.
The BGC-val toolkit was originally built to work with NEMO's extended eORCA1 grid, which is a tri-polar grid with an irregular distribution of two-dimensional latitude and longitude coordinates. However, information about the grid is supplied alongside the model data such that there is no grid requirement hard-wired into BGC-val. This means that the toolkit is capable of handling any kind of model grid, whether it be a regular grid, reversed grid, a tri-polar grid, or any other type of grid, without the need to re-interpolate the data to a common grid.
When calculating means, medians, and other metrics, the toolkit uses the grid cell area or volume to weight the results. This means that it is possible to use this toolkit to compare multiple models that use different grids without the computationally expensive and potentially lossy process of re-interpolation to a common grid. The CMIP5 datasets include grid cell area and volume. However, outside the CMIP standardised datasets, most models and observational datasets provide grid cell boundaries or corner coordinates as well as longitude and latitude cell-centred coordinates. These corners and boundaries can be used to calculate the area and volume of each grid cell. If only the cell-centred points are provided, the BGC-val toolkit is able to estimate the grid cell area and volume based on the coordinates.
2.3 Front-loaded analysis functionality
While extracting the data from file, BGC-val can apply an arbitrary predefined or user-defined mathematical Python function to the data. This means that it is straightforward to define a customised analysis function in a Python script, then to pass that function to the evaluation code, which then applies the analysis function to the dataset as the data are loaded. Firstly, this method ensures that the toolkit is not limited to a small set of predefined functions. Secondly, the end users are not required to go deep into the code repository in order to use a customised analysis function.
In its simplest form, the front-loading functionality allows a straightforward conversion of the data as they are loaded. As a basic example, it would be straightforward to add a function to convert the temperature field units from Celsius to Kelvin. The Celsius to Kelvin function would be written in a short Python script, and the script would be listed by name in the evaluation's configuration file. This custom function would be applied while loading the data, without requiring the model data to be pre-processed or for the BGC-val inner workings to be edited in depth.
Similarly, more complex analysis functions can also be front loaded in the
same way. For instance, the calculation of the global total volume of oxygen
minimum zones, the total flux of CO2 from the atmosphere to the
ocean, and the total current passing though the Drake
Passage are all relatively complex calculations which can be applied to
datasets. These functions are also already included in the toolkit in the
functions folder described in Sect. 4.3.1.
2.4 Regional masking
Similarly to the front-loading analyses described above in Sect. 2.3, BGC-val users can predefine a customised region of interest, then ignore data from outside that region. The regional definitions are supplied in advance and can be used to evaluate several models or datasets. The process of hiding data from regions that are not under investigation is known as “masking”. In addition, while the UKESM and other CMIP models are global models, there is no requirement for the model to be global in scale; regional and local models can also be investigated using BGC-val.
While BGC-val already includes many regional masks, it is straightforward to
define new masks that hide regions which are not under investigation.
Similarly to Sect. 2.3, the new masks can be defined in
advance, named, and called by name, without having to go deep into the
toolkit code. These masks can be defined in terms of the latitude, longitude,
depth, time range, or even the data. The toolkit includes several
standard masks; for example, there is a mask which allows the user to retrieve
only data in the Northern Hemisphere, called NorthernHemisphere, or
to ignore all data deeper than 10 m, called Depth_0_10m.
However, more complex masks could be created. For instance, it is feasible to make a custom mask which ignores data below the 5th percentile or above the 95th percentile. It is also possible to stack masks by applying two or more masks successively in a custom mask.
For instance, a hypothetical custom mask could mask data below a depth of 100 m, ignoring the Southern Ocean and also remove all negative values. This means that it is straightforward for users to add arbitrarily complex regional masks to the dataset. For more details, please see Sect. 4.3.2.
2.5 Interruptible
BGC-val makes regular save points during data processing such that the analysis can be interrupted and restarted without reprocessing all the data files from the beginning. This means that each analysis only needs to run once. Alternatively, it means that it is possible to evaluate on-going model simulations, without reprocessing everything every time that the evaluation is needed.
The processed data are saved as Python shelve files. Shelve files allow for any Python object, including data arrays and dictionaries, to be committed to disc. As the name suggests, shelving allows for Python objects to be stored and reloaded at a later stage. These shelve files help with the comparison of multiple models or regions, as the evaluation results can be set aside, then quickly reloaded later to be processed into a summary figure, or pushed into a human-readable data file.
2.6 Ease of use
A key goal was to make the toolkit straightforward to access, install, set up, and use. The code is accessed using a GitLab server, which is a private online graphical user interface to the version control software, Git, similar to the commercial GitHub service. This makes it straightforward for multiple users to download the code, report bugs, develop new features, and share the changes. Instructions on how to register and access the toolkit can be found below in the “Code availability” section. Once it has been cloned to your local workspace, BGC-val behaves like a standard Python package and can be installed via the “pip” interface.
More importantly, BGC-val was built such that entire evaluation suites can be run from a single human-readable configuration file. This configuration file uses the .ini configuration format and does not require any knowledge of Python or the inner workings of BGC-val. The configuration file contains all the paths to data, descriptions of the data file and model data, links to the evaluation function, Boolean switches to turn on and off various evaluation metrics, the names of the variables needed to perform an evaluation, and the paths for the output files. This makes it possible to run the entire package without having to change more than a single file. The configuration file is described in Sect. 4.1. BGC-val also summarises the results into an html document, which can be opened directly in a web browser, and evaluation figures can be extracted for publication or sharing. The summary report is described in Sect. 4.2.3. An example of the summary report is included in the Supplement.
In this section, we describe the five-stage evaluation process that the toolkit applies to model and observational data. Figure 1 summarises the evaluation process graphically.
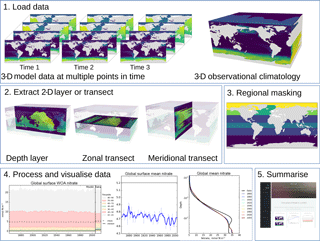
Figure 1The five stages of the evaluation process. The first stage is the loading of the model and observational data. The second stage is the extraction of a two-dimensional array. The third stage is regional masking. The fourth stage is processing and visualisation of the data. The fifth stage is the publication of an html summary report. Note that the three figures shown in the fourth stage are repeated below in Figs. 3, 4, and 6.
3.1 Load model and observational data
The first stage of the evaluation process is to load the model and observational data. The model data are typically a time series of two- or three-dimensional variables stored in one or several NetCDF files. Please note that we use the standard convention of only counting spatial dimensions. As such, any mention of dimensionality here implies an additional temporal dimension; i.e. three-dimensional model data have length, height, width, and time dimensions.
The model data can be a single NetCDF variable or some combination of several variables. For instance, in some marine biogeochemistry models, total chlorophyll concentration is calculated as the sum of many individual phytoplankton functional type chlorophyll concentrations. In all cases, BGC-val loads the model data one time step at the time, whether the NetCDFs contain one or multiple time steps.
The front-loading evaluation functions described in Sects. 2.3 and 4.3.1 are applied to each time step of the model data at this point. The resulting loaded data can be a one-, two-, or three-dimensional array. The use of an observational dataset is optional, but allows the model to be compared against historical measurements. The observational data and model data are not required to be loaded using the same function.
When loading data, BGC-val assumes that we use the NetCDF format. The NetCDF
files are opened in BGC-val with a custom Python interface,
dataset.py, in the bgcvaltools package. The dataset
class is based on the standard Python netCDF4.Dataset class. NetCDF
files are composed of two parts, the header and the data. The header
typically includes all the information needed to understand the origin of the
file, while the data contain a series of named variables. Each named
variable should (but not obligatorily) include their dimensions, units, their
long name, their data, and their mask. Furthermore, the dimensions in NetCDF
format are not restricted to regular latitude–longitude grids. Some NetCDFs
use arbitrary dimensions, such as a grid cell index, irregular grids like
NEMO's eORCA1 grid, or even triangular grid cells as in the Finite Volume
Coastal Ocean Model (FVCOM) (Chen et al., 2006).
3.2 Extract a two-dimensional slice
The second evaluation stage is the extraction of a two-dimensional variable from three-dimensional data. As shown in the second panel of Fig. 1, the two-dimensional variable can be the surface of the ocean, a depth layer parallel to the surface, an east–west transect parallel to the Equator, or a north–south transect perpendicular to the Equator. This stage is included in order to speed up the process of evaluating a model; in general, it is much quicker to evaluate a 2-D field than a 3-D field. Furthermore, the spatial and transect maps produced by the evaluation process can become visually confusing when overlaying several layers. Note that stages 2 and 3 are applied to both model and observational data (if present). This stage is unnecessary if the data loaded in the first stage are already a two-dimensional variable, such as the fractional sea ice coverage, or a one-dimensional variable, such as the Drake Passage Current.
In the case of a transect, instead of extracting along the file's internal grid, the transect is produced according to the geographic coordinates of the grid. This is done by locating points along the transect line inside the grid cells based on the grid cell corners.
3.3 Extract a specific region
Stage 3 is the masking of specific regions or depth levels from the 2-D extracted layer, as described in Sect. 2.4. Stage 3 is not needed if the variable is already a one-dimensional product, such as the total global flux of CO2. Stage 3 takes the two-dimensional slice, then converts the data into five one-dimensional arrays of equal length. These arrays represent the time, depth, latitude, longitude, and value of each data point in the data. These five 1-D arrays can be further reduced by making cuts based on any of the coordinates or even cutting according to the data.
Both stages 2 and 3 of the evaluation process reduce the number of grid cells under evaluation. This two-stage process is needed because the stage 3 masking cut can become memory intensive. As such, it is best to for the data to arrive at this stage in a reduced format. In contrast, the stage 2 process of producing a 2-D slice is a relatively computationally cheap process. This means that the overall evaluation of a model run can be done much faster.
3.4 Produce visualisations
Stage 4 is the processing of the two-dimensional datasets and the creation of visualisations of the model and observational data. Figure 1 shows three examples of the visualisations that BGC-val can produce: the time series of the spread of the data, a simple time series, and the time development of the depth profile. However, several other visualisations can also be produced: for instance, the point-to-point comparisons of model data against observational data and a comparison of the same measurement between different regions, times or models, or scenarios.
Which visualisations are produced depends on which evaluation switches are turned on, but also a range of other factors including the dimensionality of the model dataset and the presence of an observational dataset. For instance, figures that show the time development of the depth profile require three-dimensional data. Similarly, the point-to-point comparison requires an observational dataset for the model to match against. More details on the range of plotting tools are available in Sects. 4.2.1 and 4.2.2.
Stages 1–4 are repeated for each evaluated field and for multiple models scenarios or different versions of the same model. If multiple jobs or models are requested, then comparison figures can also be created in stage 4.
3.5 Produce a summary report
The fifth stage is the automated generation of a summary report. This is an html document which shows the figures that were produced as part of stages 1–4. This document is built from html and can be hosted and shared on a web server. More details on the report are available in Sect. 4.2.3.
The directory structure of the BGC-val toolkit repository is summarised in
Fig. 2. This figure highlights a handful of the key features. We
use the standard Python nomenclature where applicable. In Python, a module
is a Python source file, which can expose classes, functions, and global
variables. A package is simply a directory containing one or more modules and
Python creates a package using the __init__.py file. The BGC-val
toolkit contains seven packages and dozens of modules.
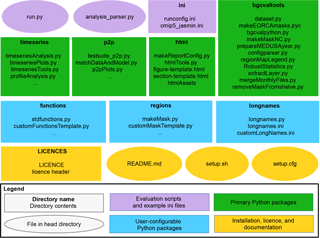
Figure 2The structure of the BGC-val repository. The principal directories are shown as rectangles, with the name of the directory in bold followed by the key files contained in that directory. Individual files in the head directory are shown with rounded corners. The evaluation scripts and the configuration directory are shown in purple. The primary Python modules are split into four directories, shown in green rectangles. The three user-configurable Python modules are shown as blue rectangles. The licence, README, and setup files are shown in yellow.
In this figure, ovals are used to show single files in the head directory,
and rectangles show folders or packages. In the top row of
Fig. 2, there are two purple ovals and a rectangle
which represent the important evaluation scripts and the example
configuration files. These files include the run.py executable
script, which is a user-friendly wrapper for the analysis_parser.py
script (also in the head directory). The analysis_parser.py file is
the principal Python file that loads the run configuration and launches the
individual analyses. The ini directory includes several example
configuration files, including the configuration files that were used to
produce the figure in this document. Note that the ini directory is
not a Python package, just a repository that holds several files. A full
description of the functionality of configuration files can be found in
Sect. 4.1.
The four main Python packages in
BGC-val are shown in green rectangles in Fig. 2. Each
of these modules has a specific purpose: the timeseries package
described in Sect. 4.2.1 performs the evaluation of the time
development of the model, the p2p package described in
Sect. 4.2.2 does an in-depth spatial comparison of a single point in
time for the model against a historical data field, and the html
package described in Sect. 4.2.3 contains all the Python functions
and html templates needed to produce the html summary report. This
bgcvaltools package contains many Python routines that perform a
range of important functions in the toolkit. These tools include, but are not
limited to, a tool to read NetCDF files, a tool to extract a specific 2-D
layer or transect, a tool to read and understand the configuration file, and
many others.
The three user-configurable packages functions, regions, and
longnames are shown in blue in Fig. 2. The
functions package, described in Sect. 4.3.1, contains
all the front-loading analysis functions described in
Sect. 2.3, which are applied in the first stage of the
evaluation process described in Sect. 3. The regions
package described in Sect. 4.3.2 contains all the masking tools
described in Sect. 2.4 and which are applied in the
third stage of the evaluation process described in Sect. 3.
The longnames package, described in Sect. 4.3.3, is a
simple tool which behaves like a look-up dictionary, allowing users to link
human-readable or “pretty” names (like “chlorophyll”) against the
internal code names or shorthand (like “chl”). The pretty names are used in
several places, notably in the figure titles and legends and on the html report.
The licences directory, the setup configuration files and the
README.md are all in the main directory of the folder, as shown in
yellow in Fig. 2. The licences directory contains
information about the Revised Berkeley Software Distribution (BSD) three-clause
licence. The README.md file contains specific details on how to
install, set up, and run the code. The setup.py and
setup.cfg files are used to install the BGC-val toolkit.
4.1 The configuration file
The configuration file is central to the running of BGC-val and contains all
the details needed to evaluate a simulation. This includes the file path of
the input model files, the user's choice of analysis regions, layers, and
functions, the names of the dimensions in the model and observational files,
the final output paths, and many other settings. All settings and
configuration choices are recorded in an single file using the .ini
format. Several example configuration files can also be found in the
ini directory. Each BGC-val configuration
file is composed of three parts: an active keys section, a list of evaluation
sections, and a global section. Each of these parts is described below.
The tools that parse the configuration file are in the
configparser.py module in the bgcvaltools package.
These tools interpret the configuration file and
use them to direct the evaluation.
Please note that we use the standard .ini format
nomenclature while describing configuration files.
In this, [Sections] are denoted with square brackets,
each option is separated from its value by a colon, “:”,
and the semi-colon “;” is the comment syntax in .ini format.
4.1.1 Active keys section
The active keys section should be the first section of any BGC-val configuration file. This section consists solely of a list of Boolean switches, one Boolean for each field that the user wants to evaluate.
[ActiveKeys] Chlorophyll : True A : False ; B : True
To reiterate the ini nomenclature, in this example
ActiveKeys is the section name,
and Chlorophyll, A, and B are options.
The values associated with these options are the Booleans
True, False, and True.
Option B is commented out and will be ignored by BGC-val.
In the [ActiveKeys] section, only options whose values are set to True are active.
False Boolean values and commented lines are not evaluated by BGC-val.
In this example, the Chlorophyll evaluation is active,
but both options A and B are switched off.
4.1.2 Individual evaluation sections
Each True Boolean option in the [ActiveKeys] section needs
an associated [Section] with the same name as the option in
the [ActiveKeys] section. The following is an example of an evaluation
section for chlorophyll in the HadGEM2-ES model.
[Chlorophyll] name : Chlorophyll units : mg C/m^3 ; The model name and paths model : HadGEM2-ES modelFiles : /Path/*.nc modelgrid : CMIP5-HadGEM2-ES gridFile : /Path/grid_file.nc ; Model coordinates/dimension names model_t : time model_cal : auto model_z : lev model_lat : lat model_lon : lon ; Data and conversion model_vars : chl model_convert : multiplyBy model_convert_factor : 1e6 dimensions : 3 ; Layers and Regions layers : Surface 100m regions : Global SouthernOcean
The name and units options are descriptive only; they are
shown on the figures and in the html report, but do not influence the
calculations. This is set up so that the name associated with the analysis
may be different to the name of the fields being loaded. Similarly, while
NetCDF files often have units associated with each field, they may not match
the units after the user has applied an evaluation function. For this reason,
the final units after any transformation must be supplied by the user. In the
example shown here, HadGEM2-ES correctly used the CMIP5 standard units for
chlorophyll concentration, kg m−3. However, we prefer to view
chlorophyll in units of mg m−3.
The model option is typically set in the Global section,
described below in Sect. 4.1.3, but it can be set here as well.
The modelFiles option is the path that BGC-val should use to locate
the model data files on local storage. The modelFiles option can
point directly at a single NetCDF file or can point to many files using
wild cards (*, ?). The file finder uses the standard Python
package glob, so wild cards must be compatible with that package.
Additional nuances can be added to the file path parser using the
placeholders $MODEL, $SCENARIO, $JOBID,
$NAME, and $USERNAME. These placeholders are replaced with
the appropriate global setting as they are read by the configparser
package. The global settings are described below in Sect. 4.1.3.
For instance, if the configuration file is set to iterate over several
models, then the $MODEL placeholder will be replaced by the model
name currently being evaluated.
The gridFile option allows BGC-val to locate the grid description
file. The grid description file is a crucial requirement for BGC-val, as it
provides important data about the model mask, the grid cell area, and the grid
cell volume. Minimally, the grid file should be a NetCDF which contains the
following information about the model grid: the cell-centred coordinates for
longitude, latitude, and depth, and these fields should use the same
coordinate system as the field currently being evaluated. In addition, the
land mask should be recorded in the grid description NetCDF in a field called
tmask, the cell area should be in a field called area, and
the cell volume should be recorded in a field labelled pvol. BGC-val
includes the meshgridmaker module in the bgcvaltools
package and the function makeGridFile from that module can be used
to produce a grid file. The meshgridmaker module can also be used to
calculate the cross-sectional area of an ocean transect, which is used in
several flux metrics such as the Drake Passage Current or the Atlantic
meridional overturning circulation.
Certain models use more than one grid to describe the ocean; for instance,
NEMO uses a U grid, a V grid, a W grid, and a T grid. In that case, care
needs to be taken to ensure that the grid file provided matches the data. The
name of the grid can be set with the modelgrid option.
The names of the coordinate fields in the NetCDF need to be provided here.
They are model_t for the time and model_cal for the model
calendar. Any NetCDF calendar option (360_day, 365_day, standard,
Gregorian, etc.) is also available using the model_cal
option; however, the code will preferentially use the calendar included in
standard NetCDF files. For more details, see the num2date function
of the netCDF4 Python package
(https://unidata.github.io/netcdf4-python/, last access: 5 October 2018). The depth, latitude, and longitude field names are passed to
BGC-val via the model_z, model_lat, and
model_lon options.
The model_vars option tells BGC-val the names of the model fields
that we are interested in. In this example, the CMIP5 HadGEM2-ES chlorophyll
field is stored in the NetCDF under the field name chl. As already mentioned, HadGEM2-ES used the CMIP5 standard units for chlorophyll
concentration, kg m−3, but we prefer to view chlorophyll in units of
mg m−3. As such, we load the chlorophyll field using the conversion
function multiplyBy and give it the argument 1e6 with the
model_convert_factor option. More details are available below in
Sect. 4.3.1 and in the README.md file.
BGC-val uses the coordinates provided here to extract the layers requested in
the layers option from the data loaded by the function in the
model_convert option. In this example that would be the surface and
the 100 m depth layer. For the time series and profile analyses, the
layer slicing is applied in the DataLoader class in the
timeseriesTools module of the timeseries package. For the
point-to-point analyses, the layer slicing is applied in the
matchDataAndModel class in the matchDataAndModel module of
the p2p package.
Once the 2-D field has been extracted, BGC-val masks the data outside the
regions requested in the regions option. In this example, that is
the Global and the SouthernOcean regions. These two regions
are defined in the regions package in the makeMask module.
This process is described below in Sect. 4.3.2.
The dimensions option tells BGC-val what the dimensionality of the
variable will be after it is loaded, but before it is masked or sliced. The
dimensionality of the loaded variable affects how the final results are
plotted. For instance, one-dimensional variables such as the global total
primary production or the total Northern Hemisphere ice extent cannot be
plotted with a depth profile or with a spatial component. Similarly,
two-dimensional variables such as the air–sea flux of CO2 or the mixed
layer depth should not be plotted as a depth profile, but can be plotted with
percentile distributions. Three-dimensional variables such as the temperature
and salinity fields, the nutrient concentrations, and the biogeochemical
advected tracers are plotted with time series, depth profile, and percentile
distributions. If any specific types of plots are possible but not wanted,
they can be switched off using one of the following options.
makeTS : True makeProfiles : False makeP2P : True
The makeTS option controls the time series plots,
the makeProfiles option controls the profile plots,
and the makeP2P option controls the point-to-point evaluation plots.
These options can be set for each active keys section,
or they can be set in the global section, as described below.
In the case of the HadGEM2-ES chlorophyll section, shown in this example, the
absence of an observational data file means that some evaluation figures will
have blank areas, and others figures will not be made at all. For instance,
it is impossible to produce a point-to-point comparison plot without both
model and observational data files. The evaluation of [Chlorophyll]
could be expanded by mirroring the model's coordinate and convert fields with
a similar set of data coordinates and convert functions for an observational
dataset.
4.1.3 Global section
The [Global] section of the configuration file can be used to set
default behaviour which is common to many evaluation sections. This is
because the evaluation sections of the configuration file often use the same
option and values in several sections. As an example, the names that a model
uses for its coordinates are typically the same between fields; i.e. a
chlorophyll data file will use the same name for the latitude coordinate as
the nitrate data file from the same model. Setting default analysis settings
in the [Global] section ensures that they do not have to be repeated
in each evaluation section. As an example, the following is a small part of a
global settings section.
[Global] model : ModelName model_lat : Latitude
These values are now the defaults, and individual evaluation sections of this
configuration file no longer require the model or
model_lat options. However, note that local settings override the
global settings. Note that certain options such as name or
units cannot be set to a default value.
The global section also includes some options that are not present in the individual field sections. For instance, each configuration file can only produce a single output report, so all the configuration details regarding the html report are kept in the global section.
[Global] makeComp : True makeReport : True reportdir : reports/HadGEM2-ES_chl
The makeComp is a Boolean flag to turn on the comparison of
multiple jobs, models, or scenarios. The makeReport is a Boolean flag
which turns on the global report making and reportdir is the path
for the html report.
The global options jobID, year, model, and
scenario can be set to a single value or can be set to multiple
values (separated by a space character) by swapping them with the options
jobIDs, years, models, or scenarios. For
instance, if multiple models were requested, then swap
[Global] model : ModelName1
with the following.
[Global] models : ModelName1 ModelName2
For the sake of the clarity of the final report, we recommend only setting one of these options with multiple values at one time. The comparison reports are clearest when grouped according to a single setting; i.e. please do not try to compare too many different models, scenarios, and job IDs at the same time.
The [Global] section also holds the paths to the location on disc
where the processed data files and the output images are to be saved.
The images are saved to the paths set with the following global options:
images_ts, images_pro, images_p2p, and images_comp
for the time series, profiles, point-to-point, and comparison figures, respectively.
Similarly, the post-processed data files are saved to the paths set with the following global options:
postproc_ts, postproc_pro, and postproc_p2p
for the time series, profiles, and point-to-point-processed data files, respectively.
As described above, the global fields jobID, year,
model, and scenario can be used as placeholders in file
paths. Following the bash shell grammar, the placeholders are marked as all
capitals with a leading $ sign. For instance, the output directory for the
time series images could be set to the following.
[Global] images_ts : images/$MODEL/$NAME
$MODEL and $NAME are placeholders for the model
name string and the name of the field being evaluated. In the example in
Sect. 4.1.2 above, the images_ts path would
become images/HadGEM2-ES/Chlorophyll. Similarly, the
basedir_model and basedir_obs global options can be used
to fill the placeholders $BASEDIR_MODEL and
$BASEDIR_OBS such that the base directory for models or
observational data does not need to be repeated in every section.
A full list of the contents of a global section can be found in the
README.md file. Also, several example configuration files are
available in the ini directory.
4.2 Primary Python packages
In this section, we describe the important packages that are shown in green
in Fig. 2. The timeseries package is described
in Sect. 4.2.1, the p2p package is described in
Sect. 4.2.2, and the html package is described in
Sect. 4.2.3. All the figures in Sect. 4.2 were
produced on the JASMIN computational resource using the example
configuration file ini/HadGEM2-ES_no3_cmip5_jasmin.ini, and the
html summary report associated with that configuration file is available in
the Supplement.
Outside the three main packages described below, the bgcvaltools
package contains many Python routines that perform a range of important
functions. These tools include a tool to read NetCDF files
dataset.py, a tool to extract a specific 2-D layer or transect
extractLayer.py, and a tool to read and understand the configuration
file, configparser.py. There is a wide and diverse selection of
tools in this directory: some of them are used regularly by the toolkit, and
some are only used in specific circumstances. More details are available in
the README.md file, and each individual module in the
bgcvaltools is sufficiently documented that its role in the toolkit
is clear.
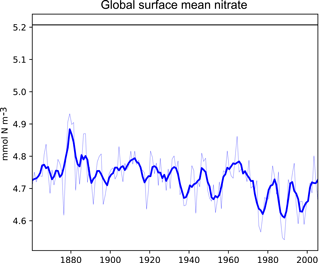
Figure 3A plot produced by the time series package. This figure shows the time development of a single metric, in this case the global surface mean nitrate in HadGEM2-ES in the historical simulation. It also shows the 5-year moving average of the metric.
4.2.1 Time series tools
This timeseries package is a set of Python tools that produces
figures showing the time development of the model. These tools manage the
extraction of data from NetCDF files, the calculation of a range of metrics
or indices, the storing and loading of processed data, and the production of
figures illustrating these metrics.
Firstly, the time development of any combination of depth layer and region
can be investigated with these tools. The spatial regions can be taken
from the predefined list or a custom region can be created. The predefined
regions are listed in the regions directory of the BGC-val. Many
metrics are available including, mean, median, minimum, maximum, and all
percentiles divisible by 10 (10th percentile, 20th percentile, etc.).
Furthermore, any user-defined custom function can also be included as a
custom function, for instance the calculation of global total integrated
primary production or the total flux through the Drake Passage.
The time series tools produce three types of analysis plots. Examples of these three types of figures are shown in Figs. 3, 4, and 5. All three examples use annual averages of the nitrate (CMIP5 name: no3) in the surface layer of the global ocean in the HadGEM2-ES model historical scenario, ensemble member r1i1p1.
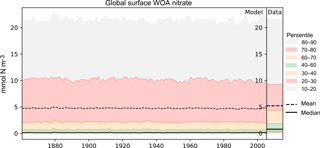
Figure 4A plot produced by the time series package. The figure shows the time developers of many metrics at once: the mean, median, and several percentile ranges of the observational data and the model data. In this case, the model data are the global surface mean nitrate in HadGEM2-ES in the historical simulation.
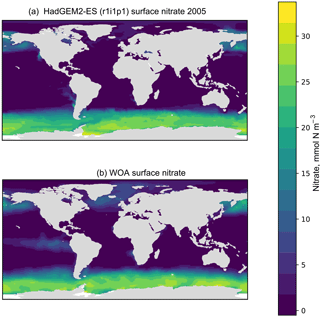
Figure 5A plot produced by the time series package. The figure shows the spatial distribution of the model (a) and the observational dataset (b). In this case, the model data are the global surface mean nitrate in HadGEM2-ES in the historical simulation.
Figure 3 shows the time development of a single variable: the mean of the nitrate in the surface layer over the entire global ocean. This figure shows the annual mean of the HadGEM2-ES model's nitrate as a thin blue line, the 5-year moving average of the HadGEM2-ES model's nitrate as a thick blue line, and the World Ocean Atlas (WOA) data (Garcia et al., 2013b) as a flat black line. The WOA data used here are an annual average climatological dataset and hence do not have a time component. This figure highlights the fact that the model simulates a decrease in the mean surface nitrate over the course of the 20th century.
Figure 4 shows an example of a percentile range plot, which shows the time development of the spatial distribution of the model data, including the mean and median, and coloured bands to indicate the 10–20, 20–30, 40–60, 60–70, and 70–80 percentile bands. This kind of plot also shows the percentile distribution of the spatial distribution of the observational data in a column on the right-hand side. Figure 4 shows the behaviour of nitrate in the surface layer over the entire global ocean, in the HadGEM2-ES model historical scenario, ensemble member r1i1p1. This type of plot is produced when the data have two or three dimensions but cannot be produced for one-dimensional model datasets. The percentile figures can be produced for any layer and spatial region and these metrics are all area weighted. For all three kinds of time series figures, a real dataset can be added, although it is not possible to include the time development of the observational dataset at this stage.
The time series package also produces a figure showing the spatial distribution of the model and observational data. Figure 5 shows an example of such a figure, in which panel (a) shows the spatial distribution of the final time step of the model, and panel (b) shows the spatial distribution of the observational dataset. It is possible to plot data for any layer for any region. These spatial distributions are made using the Plate Carré projection, and the projections are set to focus on the region in question. Figure 5 highlights the fact that the HadGEM2-ES model failed to capture the high nitrate seen in the observational data in the equatorial Pacific.
BGC-val can also produce several figures showing the time development of the
model datasets over their entire water columns. The profile modules are
stored in the timeseries package, as the time series and profiles
figures share many of the same underlying methods.
Figures 6, 7, and 8 are
examples of three profile plots showing the time development over the water
column of the global mean nitrate in the HadGEM2-ES model historical
simulation, ensemble member r1i1p1. These plots can only be produced
when the data have three dimensions. These plots can be made for any region from
the predefined list or for custom regions.
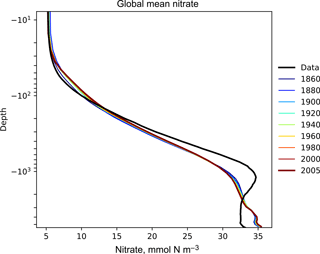
Figure 6The time development of the global mean dissolved nitrate over a range of depths. This figure shows the HadGEM2-ES global mean nitrate over the entire water column; each model year is included as a coloured line, and the annual mean of the World Ocean Atlas nitrate climatology dataset is shown as a black line.
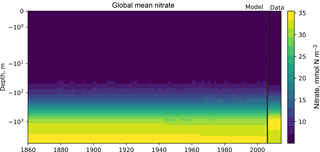
Figure 7The time development of the global mean dissolved nitrate over a range of depths. The figure shows the same data as Fig. 6, but as a Hovmöller time series plot. The annual mean of the World Ocean Atlas nitrate climatology dataset is shown as a column on the right-hand side of the figure.
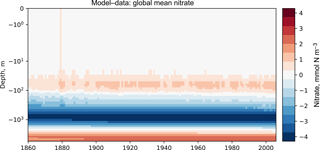
Figure 6 shows the time development of the depth profile of the model and observational data. The x axis shows the value, in this case the nitrate concentration in mmol N m−3 and the y axis shows depths. These types of plots always show the first and last time slice of the model, then a subset of the other years is also shown. Each year is assigned a different colour, with the colour scale shown in the right-hand side legend. If available, the observational data are shown as a black line. This figure shows the annual mean of the World Ocean Atlas nitrate climatology dataset as a black line.
Figures 7 and 8 are both Hovmöller diagrams (Hovmöller, 1949) and they show the depth profile over time for model and observational data. Figure 7 shows the model and the observational data side by side, and Fig. 8 shows the difference between the model data and the observational data. The Hovmöller difference diagrams are only made when an observation dataset is supplied. There appears to be a peak in the difference between the model and the observations over the entire water column in the year 1880. This does not appear to be a fault in the model, but simply a brief period during which the difference was slightly larger than zero. This peak in the mean is also visible in the global mean surface nitrate in Fig. 3, but is not visible in the percentile distribution of the surface nitrate in Fig. 4.
Figures 6, 7, and 8 all show that the model data match the observational nitrate near the surface, but diverge at depth. The model underestimates the peak in the global mean of the observational nitrate at a depth of approximately 1000 m and then overestimates the observed nitrate below 2000 m. Also, the model does not show much interannual variability in the structure of the global annual average nitrate over the 145-year simulation. However, it is unclear from the WOA annual average whether we should expect any variability from the model over this time range.
In the timeseries package, there is also a set of tools for
comparing the time series development between multiple versions of the same
metric. This is effectively the same as plotting several versions of
Fig. 3 on the same axes. This kind of diagram can be useful to
compare the same measurement between different regions, depth layers, or
different members of a model's ensemble. However, these plots can also be
used to compare multiple models. Several example figures are included in
Sect. 5.
4.2.2 Point-to-point model–data comparison tools
In addition to the time series evaluation, BGC-val can perform a direct point-to-point comparison of model against data. The point-to-point tools here are based on the work by de Mora et al. (2013). In that work, we demonstrated that using point-to-point analysis is more representative of a real marine dataset than comparing the bulk mean of the model to the bulk mean of the data. The method involves matching the model data to the closest corresponding observational measurement, then hiding all model points which do not have a corresponding observational measurement and vice versa. The point-to-point methodology means that the model and observational data have not be re-interpolated to a common grid: they both retain their original grid description.
Figures 9, 10, and 11 show examples of the figures made by the point-to-point package. In all three figures, the model data are the global surface nitrate in HadGEM2-ES in the historical simulation in ensemble member r1i1p1 in the year 2000. The observational data come from the nitrate dataset in the World Ocean Atlas (Garcia et al., 2013b).
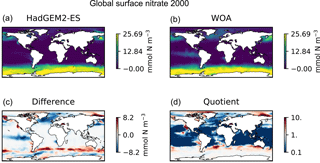
Figure 9Four spatial distributions showing the model (a), the observational data (b), the difference between them (c), and their quotient (d). The model data in these plots are the global surface nitrate in HadGEM2-ES in the historical simulation in ensemble member r1i1p1 in the year 2000. The observational data come from the annual nitrate dataset in the World Ocean Atlas.
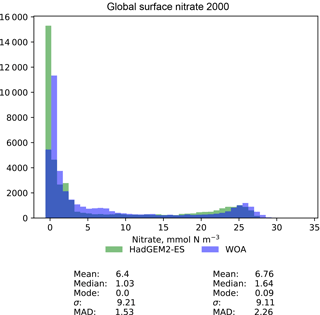
Figure 10A pair of histograms showing the model (green) and the observational data (blue), as well as some metrics of the distribution shape. The model data are the global surface nitrate in HadGEM2-ES in the historical simulation in the year 2000. The observational data come from the nitrate dataset in the World Ocean Atlas. The metrics are the mean, median, the mode, the standard deviation, σ, and the median absolute deviation (MAD).
Figure 9 is a group of four spatial distributions comparing the model and observational datasets. The map in Fig. 9a is the model, Fig. 9b is the observations, Fig. 9c is the difference between the model and observations (model minus observational data), and Fig. 9d is the quotient (model over observational data). This example shows the comparison at the ocean surface, but these tools also allow for longitudinal or latitudinal transect comparisons or a spatial distribution along a specific depth level. This figure shows that the year 2000 of the HadGEM2-ES model reproduces the large-scale spatial patterns seen in the observational dataset. The model has significantly higher nitrate that the observational climatology in the Southern Ocean, the North Pacific, and the equatorial regions and has significantly lower nitrate in the Arctic regions. A discrepancy in the spatial extent of the high nitrate in the Southern Ocean is shown clearly in the difference panel of this figure. The quotient panel of this figure also shows that model underestimates the low-nitrate regions around the tropical waters.
Figure 10 is a pair of histograms showing the same model and observational data as in Fig. 9. This figure also shows some measures of the central tendency (mean, median, mode) and measures of the deviation (standard deviation and median absolute deviation) for both model and data. These histograms confirm that the model underestimates the nitrate concentration in the low-nitrate region, which covers a significant region and is the mode of the WOA dataset.
Figure 11 shows the distribution of the model and the observational data with the model data along the x axis and the observation data along the y axis. The 1:1 line is shown as a dashed line; the model overestimates the observation to the right of this line and underestimates it to the left of this line. A linear regression is shown as a full line, with the slope, intersect, P value, correlation, and number of data points shown on the right-hand side of the figure. In this example, the linear regression is very close to the 1:1 line, and the bulk of the data is close to a good fit. While the model reproduces the distribution of observational data at low values and high nitrate concentrations, the model overestimates more than half of the nitrate observations between 10 and 20 mmol N m−3.
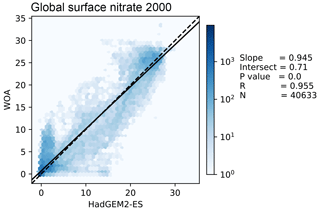
Figure 11This figure shows the distribution of the model and the observational data with the model data along the x axis and the observation data along the y axis. The model data are the global surface nitrate in HadGEM2-ES in the historical simulation in the year 2000. The observational data come from the nitrate dataset in the World Ocean Atlas. The 1:1 line is shown as a dashed line. A linear regression is shown as a full line, with the slope, intersect, P value, correlation, and number of data points shown on the right-hand side.
4.2.3 HTML report
The html package of the BGC-val toolkit contains all the tools
needed to produce a report summarising the output of the time series, the
profile, and the point-to-point packages. The principal file in this package
is the makeReportConfig module, which produces an html document
according to the settings of the configuration file. Using the configuration
file, the report maker finds all the images and uses several template files
to stitch together the individual sections of the report. The html is based
on a template taken from https://html5up.net/ (last access: 5 October 2018), used under the Creative Commons Attribution 3.0 License.
An example of the HTML report is available in the Supplement.
This report shows the output of the example configuration file:
ini/HadGEM2-ES_no3_cmip5_jasmin.ini.
In order to access this report, please download and unzip the files,
then export them to a local copy before opening the index.html file
in a browser of your choice.
4.3 User-configurable Python packages
In this section, we look at the code behind the extensive customisability of
BGC-val: the functions, regions, and longnames
packages shown in blue in Fig. 2 are described in this
section. The functions package is described in
Sect. 4.3.1, the regions package is described in
Sect. 4.3.2, and the longnames package is described in
Sect. 4.3.3.
4.3.1 Functions
The functions package is a significant contributor of the flexibility of BGC-val. This package allows any operation to be applied to a dataset as the data are loaded. In most cases, the conversion is one of the standard functions such as multiply or divide by some arbitrary number, add a constant value to the variable, or simply just load the data as is with no conversion. However, this package can also be used to perform complex data processing.
The data_convert and model_convert options in the
configuration file allow BGC-val to determine which function to apply to the
model or observational data as they are loaded. There is no default function, so
to simply load the data into the file, the standard function
NoChange should be specified in the data_convert or
model_convert options.
As an example of the structure of a basic function, we look at a simplified
version of the multiplyBy function in the stdfunctions
module of the functions package.
def multiplyBy(nc,keys, **kwargs): f = float(kwargs['factor']) return nc.variables[keys[0]][:]*f
After declaring the function name and arguments in the first line, this
function loads the factor from the keyword arguments
(kwargs) and parses it into the single precision floating format in
the second line. In the third line, this function loads the first item in the
keys list from the NetCDF dataset nc, then multiplies that
data by the factor f, and returns the product. The path to the
NetCDF file, the choice of function, the list of keys, and the factor are all
provided by the configuration file.
All functions need to be called with the same arguments: nc is a
NetCDF file opened by the dataset module from the
bgcvaltools package. The keys argument is a list of strings
which represent the names of fields in a NetCDF file, and the optional
kwargs argument is used to pass any extra information that is needed
(such as a factor or addend).
The keyword arguments which are passed to the function must be preceded by
the text model_convert_ or data_convert_ strings in the
configuration file. In the example above, the “factor” was written in the
configuration file as
model_convert_factor : 1e6
but it was loaded in the multiplyBy function as kwargs['factor'].
Some evaluation metrics require multiple variables to be loaded at once and
combined together. The stdfunctions module of the functions
package contains a few such medium-complexity operations, such as “sum”, which
returns the sum of the fields in the keys list. The
'divide' function returns the quotient of the first key over the
second key from the keys list.
More complex functions can be implemented as well, for instance depth
integration, global totals, or the flux through a certain cross section.
There are several examples of complex functions in the functions folder.
Note that some of these functions can change the dimensionality of the data,
and caution needs to be taken to ensure that the dimensions option
in the configuration file matches the dimensions of the output of this
function.
4.3.2 Regions
Similarly to the functions package described above, the regions package
allows for expanded flexibility in the evaluation of models. The term
“region” here is a portmanteau for any selection of data based on their
coordinates or values. Typically, these are spatial regional cuts, such as
“Northern Hemisphere”, but the masking is not limited to spatial regions.
For instance, the regions package can also be used to remove
negative values and to remove zero, NaN, or inf values.
As an example of the structure of a basic regional mask, we look at the
SouthHemisphere region in the
makeMask module of the regions package.
def SouthHemisphere(
name,region,
xt,xz,xy,xx,xd):
a = np.ma.masked_where(xy>0.,xd)
return a.mask
The Python standard package NumPy has been imported as
np. Each regional masking function has access to the following
fields: name, the name of the data; region, the name of the
regional cut; xt, a one-dimensional array of the dataset times;
xz, a one-dimensional array of the dataset depths; xy, a
one-dimensional array of the dataset latitudes; xx, a
one-dimensional array of the dataset longitudes; and xd, a
one-dimensional array of the data. The second to last line creates a masked
array of the data array which is masked in all the places where the latitude
coordinate is greater than zero (i.e. the Northern Hemisphere). The final line
returns the mask for this array. All region extraction functions return a
NumPy mask array. In Python, NumPy masks are an array of Booleans in which
“true”
is masked.
Many regions are already defined in the file regions/makeMask.py,
but it is straightforward to add a new region using the template file
regions/customMaskTemplate.py. To do this, make a copy of the
regions/customMaskTemplate.py file in the regions
directory, rename the function and file to your mask name, and add whatever
cuts are required. BGC-val will be able to locate your region, provided that
the region name matches the Python function and the region in your
configuration file.
4.3.3 Long names
In the Python source code, objects are often abbreviated or labelled with shorthand, and spaces and hyphens are not acceptable in object names. This means that the internal name of a model, dataset, field, layer, or region is not usually the same in the text that we want to appear in public plots. For this reason, the long name package has a dictionary of common terms with their abbreviated name linked to a “pretty” name. The dictionary has definitions for each model, scenario, dataset, object, mask, cut, region, field, and other pythonic object used in BGC-val. These pretty names are used when preparing outwards-facing graphics and html pages such that the name of an object in the configuration file is not a source of confusion.
This package uses the standard configuration (ini) format for the dictionary.
The custom long names configuration file is simply a long list of
short names as the option and long names as the value.
For example, the longnames.ini includes the following lines.
no3 : Nitrate chl : Chlorophyll
This means that we can label nitrate internally as “no3” as the evaluation
name in our configuration file, but when it appears in plots, it will be
shown as “nitrate”. Also note that the options are not case sensitive, but
the values are case sensitive. While the default long name list is already
relatively extensive, users can add their own long names to the
longnames/customLongNames.ini file.
In this section, we show some example figures of the intercomparison of
several CMIP5 models. These examples were produced using CMIP5 data on the
JASMIN data processing facility
(http://www.jasmin.ac.uk, last access: 5 October 2018),
and the configuration file used to produce these is supplied in the BGC-val
Git repository under the name cmip5_rcp85_jasmin.ini in the
ini directory. The examples that we show here are the Atlantic
meridional overturning circulation in Fig. 12, the Antarctic
circumpolar current in Fig. 13, the total annual air to sea flux
of CO2 in Fig. 14, the total annual marine
primary production in Fig. 15, and the global mean surface
chlorophyll in Fig. 16. All five figures here show the 5-year
moving average instead of the monthly or annual time resolution of the field
in order to improve clarity. The 5-year window moving average is
calculated using the mean of 2.5 years on either side of a central point.
This means that the start and end points of the time series are the mean of
only 2.5 years.
Table 1 shows the observational measurement for the multi-model mean of the years 1975–2000 in the historical scenario, the multi-model mean of the years 2075–2100 under the RCP8.5 scenario, and the percentage of change between 2075–2100 and 1975–2000 for all five fields.
These examples compare a subset of the CMIP5 models in the historical time
range and RCP8.5 scenario in the ensemble member r1i1p1. The historical and
RCP8.5 simulations are aligned such that the historical simulation links to the
RCP scenario at the year 2005. This was done using the
jasmin_cmip5_linking.py module in the bgcvaltools
package.
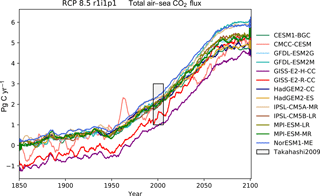
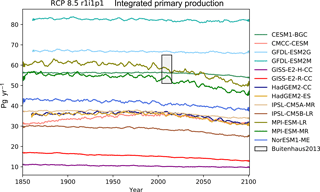
The CMIP5 models shown in these figures are CESM1-BGC, CMCC-CESM, GFDL-ESM2G, GFDL-ESM2M, GISS-E2-H-CC, GISS-E2-R-CC, HadGEM2-CC, HadGEM2-ES, IPSL-CM5A-MR, IPSL-CM5B-LR, MPI-ESM-LR, MPI-ESM-MR, and NorESM1-ME. This report does not include all CMIP5 models, but rather a small number of examples of marine circulation and biogeochemistry metrics over the historical and RCP8.5 scenarios. The selection criterion was that the model was required to have biogeochemical datasets in the British Atmospheric Data Centre (BADC) archive of the CMIP5 data. The BADC is a UK mirror of the CMIP5 data archive, which is managed by the Centre for Environmental Data Analysis (CEDA), and this archive is accessible from the JASMIN data processing facility. We also required the r1i1p1 job identifier and the “latest” model run tag.
Using these tools, we uncovered a previously undetected error in the HadGEM2-ES RCP8.5 r1i1p1 simulation. The HadGEM2-ES RC8.5 r1i1pi simulation contains 2 years in which the annual mean data were produced without all 12 months. This made the two erroneous years differ significantly from the other years in our time series plots. After informing the HadGEM2-ES project manager, we were advised to substitute the r2i1p1 simulation in place of the HadGEM2-ES r1i1p1 simulation.
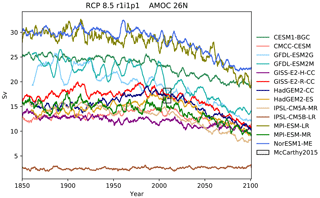
Figure 12The Atlantic meridional overturning circulation at 26∘ N in a subset of CMIP5 models. Each model is shown as a full line, and the historical measurement is shown as a grey area. The model data are a 5-year moving average.
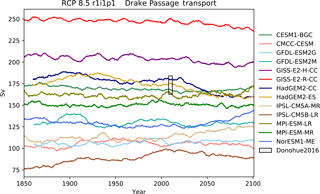
Figure 13The Drake Passage Current. Each model is shown as a full line, and the historical measurement is shown as a grey area. The model data are a 5-year moving average.
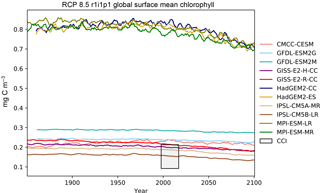
Figure 16The global mean chlorophyll concentration for the surface layer of a range of CMIP5 models.
Table 1Summary table showing the multi-model mean and standard deviation of the five fields. After the field and units columns, the observational range, measurement uncertainty, and reference are shown. The fifth column shows the multi-model mean of years 1975–2000 and the standard deviation (σ) in the historical simulation. The sixth column shows the multi-model mean of years 2075–2100 and the standard deviation in the RCP8.5 simulation. The final column (% Diff.) shows the percentage of difference between the first period and the second period.
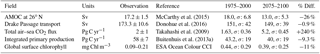
The Atlantic meridional overturning circulation (AMOC) is a major current and
consists of two parts: a northbound transport between the surface and
approximately 1200 m and a southbound transport between approximately 1200
and 3000 m (Kuhlbrodt et al., 2007). The AMOC is responsible for the
production of roughly half of the ocean's deep waters (Broecker, 1991). The
northward heat transport of the AMOC is substantial and has a significant
role in the climate of the Northern Hemisphere. The strength of the
northbound AMOC in several CMIP5 models was shown in Fig. 12.35 of the IPCC
report (Collins et al., 2013). The BGC-val toolkit was able to reproduce the
AMOC analyses of the IPCC. As in the IPCC figure, Fig. 12 shows
the historical and RCP8.5 projections of the AMOC produced by BGC-val.
Please note that we use a different subset of CMIP5 models in this figure
relative to IPCC Fig. 12.35. The RAPID array measured the long-term mean
of the AMOC to be 17.2±1.5 Sv between 2004 and 2013
(McCarthy et al., 2015). This figure is shown as a black rectangle with a grey
background in Fig. 12. The calculation was initially based on the
methods used in the UK Met Office's internal ocean evaluation toolkit, Marine
Assess, which uses the calculation described in Kuhlbrodt et al. (2007) and
McCarthy et al. (2015). However, we have since expanded the original Marine
Assess method to be model and grid independent. This cross-sectional area for
the 26∘ N transect was calculated and saved to a NetCDF file using
the meshgridmaker module in the bgcvaltools package. The
model-specific cross-sectional area was used to calculate the maximum of the
depth-integrated cross-sectional current in the custom function
cmip5AMOC in the circulation module in the
functions package. Amongst the CMIP5 models that included a
biogeochemical component, several models overestimated the AMOC, and several
underestimated the AMOC in the historical simulation. However, nearly all
simulations predict a decline in the AMOC over the 21st century, and the
multi-model mean drops by 26 % from 18 Sv in the mean of the years
1975–2000 to 13 Sv in the mean of the years 2075–2100 under the RCP8.5
scenario.
The Antarctic Circumpolar Current (ACC) is a major current which has a
significant impact on the climate of the Southern Ocean and Antarctica. The
ACC flows eastward around Antarctica and is the dominant feature of the
circulation of the Southern Ocean. The ACC was recently measured through
the Drake Passage at 173.3±10.7 Sv (Donohue et al., 2016), making the ACC the
strongest ocean current in the world. A metric to describe the ACC is the
total volume transport through the narrow gap between South America and
Antarctica, known as the Drake Passage, shown in Fig. 13. Here,
the Drake Passage Current is calculated as the total depth-integrated current
between the South American coast and the Antarctic peninsula along a line of
constant longitude at 78∘ W. To perform this calculation in a grid-independent way, a
north–south line was drawn along 78∘ W through
each model grid cell there. As in the calculation of the AMOC, described
above, this calculation was initially based on the methods used by the Met
Office's internal ocean evaluation toolkit, Marine Assess, which uses the
calculation described in Donohue et al. (2016). Like the AMOC calculation, we
expanded the Marine Assess method to be model and grid independent. The
length of the intersecting line between this line and each grid cell along
the line was calculated, then multiplied by the thickness of the layer and
the eastbound current. These products were summed together to produce the
Drake Passage Current shown in Fig. 13. This cross-sectional
area is calculated and saved to a NetCDF file using the
meshgridmaker module in the bgcvaltools package. The
calculation was performed in the custom function cmip5DrakePassage
in the circulation module in the functions package.
Figure 13 shows a moving average with a 5-year window for
several CMIP5 models between the years 1860 and 2100 in units of Sverdrups
and the observation of 173.3±10.7 Sv from Donohue et al. (2016). Several
CMIP5 models make estimates of the Drake Passage transport within the
uncertainty of the observational measurement. The percentage of difference
between the multi-model means of 1975–2000 and 2075–2100 under the RCP8.5
scenario is a decrease of 0.9 %, even though the inter-model spread is
particularly large (70–250 Sv).
The ocean is a major sink of CO2 and absorbed approximately 27 %
of anthropogenic CO2 emissions between 2002 and 2011
(Le Quéré et al., 2013). The total air–sea flux of CO2 from the
atmosphere to the ocean is an important metric for understanding the fate of
greenhouse gases (Takahashi et al., 1997). The total global air to sea flux of
CO2 from various CMIP5 models is shown in
Fig. 14 and the observational range of
2±1 Pg C yr−1 for the year 2000 is taken from
Takahashi et al. (2009). Note that the observational data were recorded between
1970 and 2007, but scaled to the year 2000. The calculation was performed in
the custom function TotalAirSeaFluxCO2 in the AirSeaFluxCO2
module in the functions package. The historical period shows a rise
in the absorption of CO2 between 1860 and 2005, and that trend is
projected to continue into the future under the RCP8.5 scenario. The multi-model
annual mean for the years 1975–2000 was 1.63 Pg of carbon per year, but
rose by 240 % up to 5.2 Pg of carbon per year for the years 2075–2100.
The integrated primary production is the global sum of the primary production
in the ocean. Marine phytoplankton are responsible for 56±7 Pg of primary
production per year (Buitenhuis et al., 2013a), which is of similar magnitude to that of
land plants (Field et al., 2011). The total primary production is an indicator
of the strength of the base of the food chain. Changes in primary production
may indicate severe impacts of climate change (Anav et al., 2013; Chavez et al., 2011). In
order to calculate this value, we multiply the primary production from each
grid cell by the volume of that grid cell, then take the global sum over the
entire ocean. The calculation was performed in the custom function
TotalIntPP in the TotalIntPP module in the
functions package. Figure 15 shows a wide range of
behaviours for the CMIP5 models. Some models show relatively consistent
marine primary production, and some models show constant historical primary
production, followed by a decrease in primary production going into the 21st
century. One model, CMCC-CESM, even shows an increase in the
20th followed by a decrease in the 21st century. The
multi-model mean annual primary production for the years 1975–2000 was
43.2 Pg of carbon per year, but decreased by 9 % down to 39.7 Pg of
carbon per year for the years 2075–2100.
The concentration of chlorophyll in the surface of the ocean is a indicator
of the quantity of phytoplankton in the waters. The global mean surface
chlorophyll for the CMIP5 models (excluding the CESM1-BGC and NorESM1-ME models)
is shown in Fig. 16. The observational data shown in
Fig. 16 and in the chlorophyll row of Table 1
are from the ESA Ocean Colour Climate Change Initiative (CCI). The CCI data
are taken from the Ocean Colour Climate Change Initiative dataset version
3.1, available online at
http://www.esa-oceancolour-cci.org (last access: 5 October 2018). The model value for global mean surface chlorophyll was calculated
by taking the 5-year moving average of the time series of the area-weighted
mean of the surface layer for each CMIP5 model. To convert the model data
into mg Chl m−3, we used the standard function multiplyBy
with a multiplicand of 1e6 from the stdfunctions module in the
functions package. The CCI global mean surface chlorophyll ranges
from 0.09 to 0.21 mg Chl m−3 for the years 1997 to 2017. Note that
this value represents the range of monthly means and was extracted using the
ocean colour data portal
https://www.oceancolour.org/portal/ (last access: 5 October 2018). This value is taken from remote sensing satellite measurements and
does not have consistent coverage due to cloud cover and low light in the
winter in the polar regions. The multi-model area-weighted mean surface
chlorophyll for the years 1975–2000 was 0.44 mg Chl per cubic metre, but
decreased by 11 % down to 0.39 mg of chlorophyll per cubic metre for the
years 2075–2100. There appear to be two modes of behaviour in the
chlorophyll of the CMIP5 models. The first grouping seems to overestimate the
total chlorophyll and the second group is closer to the observed value from
ESA Ocean Colour CCI.
The computational cost required to perform these evaluations depends on
several factors, including the number of models being investigated, the
number of years being investigated, the size of the model grid, the number of
depth fields, the number of metrics requested, the number of regions
requested, the number of depth layers requested, the number of fields under
investigation, and the power of the computational system being used. To give
a coarse estimate of the computational cost of the tool, we ran a single
thread of BGC-val over a single model (HadGEM2-ES), for a single CMIP5 field
(NO3), over a single layer (surface), in a single region (global),
over the entire CMIP5 historical period (1850–2007) and ran the time
series, profile maker, a point-to-point comparison, and the html report maker.
We used the JASMIN sci1 processing node, and ran three iterations. The
average “wall-clock” time needed to run all evaluation metrics, produce all
plots, and make the final html report was 5 min 39 s, and the peak memory usage
(resident set size) was around 4 GB, as reported by the Linux utility
time. This individual process can be parallelised such that several
threads (usually different fields) can be run at once. Due to the time-saving
tools described in Sect. 2.5, running the same evaluation
a second time only took 4 to 5 s.
The biogeochemical evaluation toolkit, BGC-val, is a model- and grid-independent toolkit that has been built to evaluate marine biogeochemical models using a simple interface. We have presented the ideas that motivated the development of the BGC-val software framework, introduced the code structure, and shown some applications of the toolkit using model results from CMIP5.
We hope that we have successfully communicated the power and flexibility of this toolkit for the automation of marine model evaluation. This toolkit has already been deployed operationally to evaluate the spin-up phase of the marine component of the UKESM1. In the future, the authors will continue to develop and apply the toolkit outlined in this work. Furthermore, as it is straightforward to add new fields and model comparisons to BGC-val, we intend to continue to use this toolkit to compare UKESM1 and the other models submitted to CMIP6 against each other and against the CMIP5 models.
In addition, the framework that produces these figures was built to make it straightforward to load, mask, and compare model and observations, as well as share results. There are several potential expansions; for instance, it may be interesting to evaluate the production of emergent features in marine biogeochemical models based on the work of de Mora et al. (2016). Another feature which is currently under development is the production of pattern statistics diagrams, such as Taylor and target diagrams (Jolliff et al., 2009; Taylor, 2001).
While ESMValTool is a separate toolkit, many of the evaluation metrics used in BGC-val are also planned to be ported onto the ESMValTool (Poloczanska et al., 2016) platform by the authors of this paper. When ported into ESMValTool version 2, these metrics will be made available for use by the wider Earth system model evaluation community.
The BGC-val toolkit is freely available and distributed with the Berkeley Software Distribution (BSD) three-clause licence. A fully functional and documented snapshot of the BGC-val toolkit with an associated DOI address will be permanently available via the Zenodo service:
An up-to-date version will be available via our in-house GitLab server. Registration for the PML GitLab service is required at
-
http://www.pml.ac.uk/Modelling_at_PML/Access_Code (last access: 5 October 2018).
The up-to-date code is available to registered GitLab users at
-
https://gitlab.ecosystem-modelling.pml.ac.uk/BGC-val-users/bgc-val (last access: 5 October 2018).
Specific and up-to-date details on how to install, set up, and run the code
can be found in the README.md file in the code repository. However,
in this appendix, we present a bare-bones guide on how to use the BGC-val
toolkit.
BGC-val was written to be compatible with Python 2.7 and has only been tested in a Linux environment. It requires several standard Python packages, including matplotlib, netCDF4, NumPy, and SciPy. It also requires a small number of nonstandard packages, such as the UKMO's cartopy package.
While the BGC-val code is available via the Zenodo service (de Mora et al., 2018), this is a snapshot and cannot be changed once it is published. The up-to-date versions of this repository will be available only with the GitLab service. For this reason, we recommend using the GitLab version instead of the Zenodo version. Instructions on how to register and access the toolkit can be found above in the “Code availability” section. The registration process will create a user account for you, and your account will be added to the BGC-val users group. Once registered, the repository can be cloned using the standard Git methodology.
Git clone git@gitlab.em.pml.ac.uk: BGC-val-users/bgc-val.git
However, note that this address may differ in the future.
Once cloned, the BGC-val repository can be installed using the standard Python package installer, pip.
pip install --user bgc-val-public
This will make the tools available in the user's Python working space.
To run the code, we advise users to make a copy of the relevant configuration
file in the ini directory. Their local copy of the configuration
file should then be edited as described in Sect. 4.1 to reflect
their local evaluation requirements.
The BGC-val toolkit is launched by the following command.
run.py configuration.ini
run.py is a simple wrapper that passes the local
configuration file as a command line argument to the main script,
analysis_parser.py.
The supplement related to this article is available online at: https://doi.org/10.5194/gmd-11-4215-2018-supplement.
LdM developed, tested, and deployed the BGC-val toolkit and wrote this paper. LdM, AY, JP, AS, TK, and CJ all contributed to the development of the scientific methodology, metrics, and graphical tools and contributed to the final paper. CJ, EP, and JIA helped develop the initial idea and supervised the subsequent developments. All authors provided critical feedback and helped shape the BGC-val toolkit, analysis, and paper.
The authors declare that they have no conflict of interest.
The software described in this paper is provided “as is” without warranty of any kind, either express or implied, including, but not limited to, the implied warranties of fitness for a purpose or the warranty of non-infringement.
We make no warranty that the software will meet your requirements, that the software will be uninterrupted, timely, secure, or error free, that the results that may be obtained from the use of the software will be effective, accurate, or reliable, that the quality of the software will meet your expectations, or that any errors in the software will be corrected.
The software and its documentation could include technical or other mistakes, inaccuracies, or typographical errors. The software or documentation here may be out of date, and the authors make no commitment to update such materials. The authors assume no responsibility for errors or omissions in the software or documentation.
In no event shall the authors be liable to you or any third parties for any special, punitive, incidental, indirect, or consequential damages of any kind or any damages whatsoever, including, without limitation, those resulting from loss of use, data, or profits, whether or not the authors have been advised of the possibility of such damages, and on any theory of liability arising out of or in connection with the use of this software.
The use of this software is done at your own discretion and risk and with agreement that you will be solely responsible for any damage to your computer system or loss of data that results from such activities. No advice or information, whether oral or written, obtained by you from the authors or from this publication website shall create any warranty for the software.
The work for this paper was funded through the National Environmental Research Council (NERC) National Capability grant to the UK Earth System Modelling project, UKESM (grant numbers NE/N018036/1 and NE/N017951/1). Andrew Yool, Till Kuhlbrodt, Colin Jones, and Ekaterina Popova were also supported by the EU Horizon 2020 Research Programme CRESCENDO project, grant agreement number 641816. AS was supported by the Met Office Hadley Centre Climate Programme funded by BEIS and Defra.
We acknowledge use of the JASMIN data processing facility, a collaborative facility supplied by the Centre for Environmental Data Analysis (CEDA) to support the data analysis requirements of the UK and European climate and Earth system modelling community, and we would like to thank the JASMIN team for their support.
We also thank the UKESM1 team and Ocean Assess/Marine Assess team of the Met Office Hadley Centre.
We acknowledge the use of the Ocean Colour Climate Change Initiative dataset,
version 3.1, European Space Agency, available online at
http://www.esa-oceancolour-cci.org (last access: 5 October 2018).
Edited by: Julia Hargreaves and Lauren Gregoire
Reviewed by: two anonymous referees
Anav, A., Friedlingstein, P., Kidston, M., Bopp, L., Ciais, P., Cox, P., Jones, C., Jung, M., Myneni, R., and Zhu, Z.: Evaluating the land and ocean components of the global carbon cycle in the CMIP5 earth system models, J. Climate, 26, 6801–6843, https://doi.org/10.1175/JCLI-D-12-00417.1, 2013. a
Appel, K. W., Gilliam, R. C., Davis, N., Zubrow, A., and Howard, S. C.: Overview of the atmospheric model evaluation tool (AMET) v1.1 for evaluating meteorological and air quality models, Environ. Modell. Softw., 26, 434–443, https://doi.org/10.1016/j.envsoft.2010.09.007, 2011. a
Azevedo, L. B., De Schryver, A. M., Hendriks, A. J., and Huijbregts, M. A.: Calcifying species sensitivity distributions for ocean acidification, Environ. Sci. Technol., 49, 1495–1500, https://doi.org/10.1021/es505485m, 2015. a
Boyd, P. W. and Trull, T. W.: Understanding the export of biogenic particles in oceanic waters: Is there consensus?, Prog. Oceanogr., 72, 276–312, https://doi.org/10.1016/j.pocean.2006.10.007, 2007. a
Broecker, W. S.: The Great Ocean Conveyor, Oceanography, 4, 79–89, https://doi.org/10.5670/oceanog.1991.07, 1991. a
Brown, P. T. and Caldeira, K.: Greater future global warming inferred from Earth's recent energy budget, Nature, 552, 45–50, https://doi.org/10.1038/nature24672, 2017. a
Buitenhuis, E. T., Li, W. K. W., Vaulot, D., Lomas, M. W., Landry, M. R., Partensky, F., Karl, D. M., Ulloa, O., Campbell, L., Jacquet, S., Lantoine, F., Chavez, F., Macias, D., Gosselin, M., and McManus, G. B.: Picophytoplankton biomass distribution in the global ocean, Earth Syst. Sci. Data, 4, 37–46, https://doi.org/10.5194/essd-4-37-2012, 2012. a
Buitenhuis, E. T., Hashioka, T., and Quéré, C. L.: Combined constraints on global ocean primary production using observations and models, Global Biogeochem. Cy., 27, 847–858, https://doi.org/10.1002/gbc.20074, 2013a. a, b
Buitenhuis, E. T., Vogt, M., Moriarty, R., Bednarṽsek, N., Doney, S. C., Leblanc, K., Le Quéré, C., Luo, Y.-W., O'Brien, C., O'Brien, T., Peloquin, J., Schiebel, R., and Swan, C.: MAREDAT: towards a world atlas of MARine Ecosystem DATa, Earth Syst. Sci. Data, 5, 227–239, https://doi.org/10.5194/essd-5-227-2013, 2013b. a
Caldeira, K. and Wickett, M. E.: Anthropogenic carbon and ocean pH, Nature, 425, 365, https://doi.org/10.1038/425365a, 2003. a
Chavez, F. P., Messié, M., and Pennington, J. T.: Marine Primary Production in Relation to Climate Variability and Change, Annu. Rev. Mar. Sci., 3, 227–260, https://doi.org/10.1146/annurev.marine.010908.163917, 2011. a
Chen, C., Beardsley, R., and Cowles, G.: An Unstructured Grid, Finite-Volume Coastal Ocean Model (FVCOM) System, Oceanography, 19, 78–89, https://doi.org/10.5670/oceanog.2006.92, 2006. a
Church, J., Clark, P., Cazenave, A., Gregory, J., Jevrejeva, S., Levermann, A., Merrifield, M., Milne, G., Nerem, R., Nunn, P., Payne, A. J., Pfeffer, W., Stammer, D., and Unnikrishnan, A. S.: Sea level change, Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, 1137–1216, https://doi.org/10.1017/CB09781107415315.026, 2013. a
Collins, M., Knutti, R., Arblaster, J., Dufresne, J.-L., Fichefet, T., Friedlingstein, P., Gao, X., Gutowski, W. J., Johns, T., Krinner, G., Shongwe, M., Tebaldi, C., Weaver, A. J., and Wehner, M.: Long-term Climate Change: Projections, Commitments and Irreversibility, Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, 1029–1136, https://doi.org/10.1017/CBO9781107415324.024, 2013. a
Cook, J., Nuccitelli, D., Green, S. A., Richardson, M., Winkler, B., Painting, R., Way, R., Jacobs, P., and Skuce, A.: Quantifying the consensus on anthropogenic global warming in the scientific literature, Environ. Res. Lett., 8, 024024, https://doi.org/10.1088/1748-9326/8/2/024024, 2013. a
de Mora, L., Butenschön, M., and Allen, J. I.: How should sparse marine in situ measurements be compared to a continuous model: an example, Geosci. Model Dev., 6, 533–548, https://doi.org/10.5194/gmd-6-533-2013, 2013. a, b
de Mora, L., Butenschön, M., and Allen, J. I.: The assessment of a global marine ecosystem model on the basis of emergent properties and ecosystem function: a case study with ERSEM, Geosci. Model Dev., 9, 59–76, https://doi.org/10.5194/gmd-9-59-2016, 2016. a, b
de Mora, L., Yool, A., Palmieri, J., Sellar, A., Kuhlbrodt, T., Popova, E., Jones, C., and Icarus Allen, J.: BGC-val: a model and grid independent python toolkit to evaluate marine biogeochemical models, https://doi.org/10.5281/zenodo.1424505, 2018. a, b
Donohue, K. A., Tracey, K. L., Watts, D. R., Chidichimo, M. P., and Chereskin, T. K.: Mean Antarctic Circumpolar Current transport measured in Drake Passage, Geophys. Res. Lett., 43, 11760–11767, https://doi.org/10.1002/2016GL070319, 2016. a, b, c, d
Dutkiewicz, S., Morris, J. J., Follows, M. J., Scott, J., Levitan, O., Dyhrman, S. T., and Berman-Frank, I.: Impact of ocean acidification on the structure of future phytoplankton communities, Nat. Clim. Change, 5, 1002–1006, https://doi.org/10.1038/nclimate2722, 2015. a, b
Eyring, V., Righi, M., Lauer, A., Evaldsson, M., Wenzel, S., Jones, C., Anav, A., Andrews, O., Cionni, I., Davin, E. L., Deser, C., Ehbrecht, C., Friedlingstein, P., Gleckler, P., Gottschaldt, K.-D., Hagemann, S., Juckes, M., Kindermann, S., Krasting, J., Kunert, D., Levine, R., Loew, A., Mäkelä, J., Martin, G., Mason, E., Phillips, A. S., Read, S., Rio, C., Roehrig, R., Senftleben, D., Sterl, A., van Ulft, L. H., Walton, J., Wang, S., and Williams, K. D.: ESMValTool (v1.0) – a community diagnostic and performance metrics tool for routine evaluation of Earth system models in CMIP, Geosci. Model Dev., 9, 1747–1802, https://doi.org/10.5194/gmd-9-1747-2016, 2016. a
Field, C. B., Behrenfeld, M. J., Randerson, J. T., Falkowski, P., Field, C. B., Behrenfeld, M. J., and Randerson, J. T.: Primary Production of the Biosphere, Integrating Terrestrial and Oceanic Components, 281, 237–240, 2011. a
Fowler, T., Gotway, J. H., Newman, K., Brown, B., Bullock, R., and Jensen, T.: Model Evaluation Tools Version 7.0 (METv7.0) User's Guide, vol. 0, https://doi.org/10.1016/j.envsoft.2011.09.008, 2018. a
Garcia, H. E., Boyer, T. P., Locarnini, R. A., Antonov, J. I., Mishonov, A. V., Baranova, O. K., Zweng, M. M., Reagan, J. R., and Johnson, D. R.: World Ocean Atlas 2013. Volume 3: dissolved oxygen, apparent oxygen utilization, and oxygen saturation, NOAA Atlas NESDIS 75, 1–27, 2013a. a
Garcia, H. E., Locarnini, R. A., Boyer, T. P., Antonov, J. I., Baranova, O. K., Zweng, M. M., Reagan, J., and Johnson, D. R.: WORLD OCEAN ATLAS 2013 Volume 4: Dissolved Inorganic Nutrients, in: World Ocean Atlas, vol. 86, p. 25, 2013b. a, b, c
Garcia-Castellanos, D. and Lombardo, U.: Poles of inaccessibility: A calculation algorithm for the remotest places on earth, Scot. Geogr. J., 123, 227–233, https://doi.org/10.1080/14702540801897809, 2007. a
Gettelman, A., Eyring, V., Fischer, C., Shiona, H., Cionni, I., Neish, M., Morgenstern, O., Wood, S. W., and Li, Z.: A community diagnostic tool for chemistry climate model validation, Geosci. Model Dev., 5, 1061–1073, https://doi.org/10.5194/gmd-5-1061-2012, 2012. a
Grant, M., Jackson, T., Chuprin, A., Sathyendranath, S., Zühlke, M., Dingle, J., Storm, T., Boettcher, M., Jackson, T., Groom, S., and Sathyendranath, S.: Ocean Colour Climate Change Initiative (OC_CCI) – Phase Two Product User Guide, Tech. Rep. 3.1.0, 2017. a
Gruber, N.: Warming up, turning sour, losing breath: ocean biogeochemistry under global change, Philosophical Transactions of the Royal Society A: Mathematical, Phys. Eng. Sci., 369, 1980–1996, https://doi.org/10.1098/rsta.2011.0003, 2011. a
Henson, S. A., Sanders, R., Madsen, E., Morris, P. J., Le Moigne, F., and Quartly, G. D.: A reduced estimate of the strength of the ocean's biological carbon pump, Geophys. Res. Lett., 38, 10–14, https://doi.org/10.1029/2011GL046735, 2011. a
Hoffman, F. M., Koven, C. D., Keppel-Aleks, G., Lawrence, D. M., Riley, W. J., Randerson, J. T., Ahlström, A., Abramowitz, G., Baldocchi, D. D., Best, M. J., Bond-Lamberty, B., De Kauwe, M. G., Denning, A. S., Desai, A. R., Eyring, V., Fisher, J. B., Fisher, R. A., Gleckler, P. J., Huang, M., Hugelius, G., Jain, A. K., Kiang, N. Y., Kim, H., Koster, R. D., Kumar, S. V., Li, H., Luo, Y., Mao, J., McDowell, N. G., Mishra, U., Moorcroft, P. R., Pau, G. S., Ricciuto, D. M., Schaefer, K., Schwalm, C. R., Serbin, S. P., Shevliakova, E., Slater, A. G., Tang, J., Williams, M., Xia, J., Xu, C., Joseph, R., and Koch, D.: 2016 International Land Model Benchmarking (ILAMB) Workshop Report, Tech. rep., https://doi.org/10.5281/zenodo.1320830, 2017. a
Hovmöller, E.: The Trough-and-Ridge diagram, Tellus, 1, 62–66, https://doi.org/10.3402/tellusa.v1i2.8498, 1949. a
Jolliff, J. K., Kindle, J. C., Shulman, I., Penta, B., Friedrichs, M. A. M., Helber, R., and Arnone, R. A.: Summary diagrams for coupled hydrodynamic-ecosystem model skill assessment, J. Marine Syst., 76, 64–82, https://doi.org/10.1016/j.jmarsys.2008.05.014, 2009. a, b
Kuhlbrodt, T., Griesel, A., Montoya, M., Levermann, A., Hofmann, M., and Rahmstorf, S.: On the driving processes of the Atlantic meridional overturning circulation, Rev. Geophys., 45, 1–32, https://doi.org/10.1029/2004RG000166.1.INTRODUCTION, 2007. a, b
Kumar, S. V., Peters-Lidard, C. D., Santanello, J., Harrison, K., Liu, Y., and Shaw, M.: Land surface Verification Toolkit (LVT) – a generalized framework for land surface model evaluation, Geosci. Model Dev., 5, 869–886, https://doi.org/10.5194/gmd-5-869-2012, 2012. a
Kwiatkowski, L., Yool, A., Allen, J. I., Anderson, T. R., Barciela, R., Buitenhuis, E. T., Butenschön, M., Enright, C., Halloran, P. R., Le Quéré, C., de Mora, L., Racault, M.-F., Sinha, B., Totterdell, I. J., and Cox, P. M.: iMarNet: an ocean biogeochemistry model intercomparison project within a common physical ocean modelling framework, Biogeosciences, 11, 7291–7304, https://doi.org/10.5194/bg-11-7291-2014, 2014. a
Leblanc, K., Arístegui, J., Armand, L., Assmy, P., Beker, B., Bode, A., Breton, E., Cornet, V., Gibson, J., Gosselin, M.-P., Kopczynska, E., Marshall, H., Peloquin, J., Piontkovski, S., Poulton, A. J., Quéguiner, B., Schiebel, R., Shipe, R., Stefels, J., van Leeuwe, M. A., Varela, M., Widdicombe, C., and Yallop, M.: A global diatom database – abundance, biovolume and biomass in the world ocean, Earth Syst. Sci. Data, 4, 149–165, https://doi.org/10.5194/essd-4-149-2012, 2012. a
Le Quéré, C., Andres, R. J., Boden, T., Conway, T., Houghton, R. A., House, J. I., Marland, G., Peters, G. P., van der Werf, G. R., Ahlström, A., Andrew, R. M., Bopp, L., Canadell, J. G., Ciais, P., Doney, S. C., Enright, C., Friedlingstein, P., Huntingford, C., Jain, A. K., Jourdain, C., Kato, E., Keeling, R. F., Klein Goldewijk, K., Levis, S., Levy, P., Lomas, M., Poulter, B., Raupach, M. R., Schwinger, J., Sitch, S., Stocker, B. D., Viovy, N., Zaehle, S., and Zeng, N.: The global carbon budget 1959–2011, Earth Syst. Sci. Data, 5, 165–185, https://doi.org/10.5194/essd-5-165-2013, 2013. a, b, c
Locarnini, R. A., Mishonov, A. V., Antonov, J. I., Boyer, T. P., Garcia, H. E., Baranova, O. K., Zweng, M. M., Paver, C. R., Reagan, J. R., Johnson, D. R., Hamilton, M., and Seidov, D.: World Ocean Atlas 2013. Vol. 1: Temperature, edited by: Levitus, S. and Mishonov, A., NOAA Atlas NESDIS, 73, 1–40, 2013. a
Mahowald, N. M., Baker, A. R., Bergametti, G., Brooks, N., Duce, R. A., Jickells, T. D., Kubilay, N., Prospero, J. M., and Tegen, I.: Atmospheric global dust cycle and iron inputs to the ocean, Global Biogeochem. Cy., 19, GB4025, https://doi.org/10.1029/2004GB002402, 2005. a
McCarthy, G. D., Smeed, D. A., Johns, W. E., Frajka-Williams, E., Moat, B. I., Rayner, D., Baringer, M. O., Meinen, C. S., Collins, J., and Bryden, H. L.: Measuring the Atlantic Meridional Overturning Circulation at 26∘N, Prog. Oceanogr., 130, 91–111, https://doi.org/10.1016/j.pocean.2014.10.006, 2015. a, b, c
Meehl, G. A., Goddard, L., Murphy, J., Stouffer, R. J., Boer, G., Danabasoglu, G., Dixon, K., Giorgetta, M. A., Greene, A. M., Hawkins, E. D., Hegerl, G., Karoly, D., Keenlyside, N., Kimoto, M., Kirtman, B., Navarra, A., Pulwarty, R., Smith, D., Stammer, D., and Stockdale, T.: Decadal prediction: Can it be skillful?, B. Am. Meteorol. Soc., 90, 1467–1485, https://doi.org/10.1175/2009BAMS2778.1, 2009. a
Moore, G. W. K., Vage, K., Pickart, R. S., and Renfrew, I. A.: Decreasing intensity of open-ocean convection in the Greenland and Iceland seas, Nat. Clim. Change, 5, 877–882, https://doi.org/10.1038/nclimate2688, 2015. a
Moriarty, R. and O'Brien, T. D.: Distribution of mesozooplankton biomass in the global ocean, Earth Syst. Sci. Data, 5, 45–55, https://doi.org/10.5194/essd-5-45-2013, 2013. a
Moss, R. H., Edmonds, J. A., Hibbard, K. A., Manning, M. R., Rose, S. K., Van Vuuren, D. P., Carter, T. R., Emori, S., Kainuma, M., Kram, T., Meehl, G. A., Mitchell, J. F., Nakicenovic, N., Riahi, K., Smith, S. J., Stouffer, R. J., Thomson, A. M., Weyant, J. P., and Wilbanks, T. J.: The next generation of scenarios for climate change research and assessment, Nature, 463, 747–756, https://doi.org/10.1038/nature08823, 2010. a
Peloquin, J., Swan, C., Gruber, N., Vogt, M., Claustre, H., Ras, J., Uitz, J., Barlow, R., Behrenfeld, M., Bidigare, R., Dierssen, H., Ditullio, G., Fernandez, E., Gallienne, C., Gibb, S., Goericke, R., Harding, L., Head, E., Holligan, P., Hooker, S., Karl, D., Landry, M., Letelier, R., Llewellyn, C. A., Lomas, M., Lucas, M., Mannino, A., Marty, J.-C., Mitchell, B. G., Muller-Karger, F., Nelson, N., O'Brien, C., Prezelin, B., Repeta, D., Jr. Smith, W. O., Smythe-Wright, D., Stumpf, R., Subramaniam, A., Suzuki, K., Trees, C., Vernet, M., Wasmund, N., and Wright, S.: The MAREDAT global database of high performance liquid chromatography marine pigment measurements, Earth Syst. Sci. Data, 5, 109–123, https://doi.org/10.5194/essd-5-109-2013, 2013. a
Poloczanska, E. S., Burrows, M. T., Brown, C. J., García Molinos, J., Halpern, B. S., Hoegh-Guldberg, O., Kappel, C. V., Moore, P. J., Richardson, A. J., Schoeman, D. S., and Sydeman, W. J.: Responses of Marine Organisms to Climate Change across Oceans, Frontiers in Marine Science, 3, 1–21, https://doi.org/10.3389/fmars.2016.00062, 2016. a, b, c
Rhein, M., Rintoul, S., Aoki, S., Campos, E., Chambers, D., Feely, R., Gulev, S., Johnson, G., Josey, S., Kostianoy, A., Mauritzen, C., Roemmich, D., Talley, L., and Wang, F.: Observations: Ocean Pages, in: Climate Change 2013 – The Physical Science Basis, edited by Intergovernmental Panel on Climate Change, Cambridge University Press, Cambridge, 255–316, https://doi.org/10.1017/CBO9781107415324.010, 2013. a, b, c
Saux Picart, S., Butenschön, M., and Shutler, J. D.: Wavelet-based spatial comparison technique for analysing and evaluating two-dimensional geophysical model fields, Geosci. Model Dev., 5, 223–230, https://doi.org/10.5194/gmd-5-223-2012, 2012. a
Schleussner, C. F., Rogelj, J., Schaeffer, M., Lissner, T., Licker, R., Fischer, E. M., Knutti, R., Levermann, A., Frieler, K., and Hare, W.: Science and policy characteristics of the Paris Agreement temperature goal, Nat. Clim. Change, 6, 827–835, https://doi.org/10.1038/nclimate3096, 2016. a
Stocker, T. F., Qin, D., Plattner, G.-K., Tignor, M. M. B., Allen, S. K., Boschung, J., Nauels, A., Xia, Y., Bex, V., and Midgley, P. M. (Eds.): Summary for Policymakers, in: Climate Change 2013 – The Physical Science Basis, edited by Intergovernmental Panel on Climate Change, Cambridge University Press, Cambridge, vol. 1542, 1–30, https://doi.org/10.1017/CBO9781107415324.004, 2015. a
Stow, C. A., Jolliff, J., McGillicuddy, D. J., Doney, S. C., Allen, J. I., Friedrichs, M. A., Rose, K. A., and Wallhead, P.: Skill assessment for coupled biological/physical models of marine systems, J. Marine Syst., 76, 4–15, https://doi.org/10.1016/j.jmarsys.2008.03.011, 2009. a
Stramma, L., Brandt, P., Schafstall, J., Schott, F., Fischer, J., and Körtzinger, A.: Oxygen minimum zone in the North Atlantic south and east of the Cape Verde Islands, J. Geophys. Res.-Oceans, 113, C04014, https://doi.org/10.1029/2007JC004369, 2008. a
Takahashi, T., Feely, R. A., Weiss, R. F., Wanninkhof, R. H., Chipman, D. W., Sutherland, S. C., and Takahashi, T. T.: Global air-sea flux of CO2: an estimate based on measurements of sea-air pCO2 difference, P. Natl. Acad. Sci. USA, 94, 8292–8299, https://doi.org/10.1073/pnas.94.16.8292, 1997. a
Takahashi, T., Sutherland, S. C., Wanninkhof, R., Sweeney, C., Feely, R. A., Chipman, D. W., Hales, B., Friederich, G., Chavez, F., Sabine, C., Watson, A., Bakker, D. C., Schuster, U., Metzl, N., Yoshikawa-Inoue, H., Ishii, M., Midorikawa, T., Nojiri, Y., Körtzinger, A., Steinhoff, T., Hoppema, M., Olafsson, J., Arnarson, T. S., Tilbrook, B., Johannessen, T., Olsen, A., Bellerby, R., Wong, C. S., Delille, B., Bates, N. R., and de Baar, H. J.: Climatological mean and decadal change in surface ocean pCO2, and net sea-air CO2 flux over the global oceans, Deep-Sea Res. Pt. II, 56, 554–577, https://doi.org/10.1016/j.dsr2.2008.12.009, 2009. a, b, c, d
Taylor, K. E.: Summarizing multiple aspects of model performance in a single diagram, J. Geophys. Res.-Atmos., 106, 7183–7192, https://doi.org/10.1029/2000JD900719, 2001. a, b
Taylor, K. E., Stouffer, R. J., and Meehl, G. A.: A Summary of the CMIP5 Experiment Design, World, 4, 1–33, https://doi.org/10.1175/BAMS-D-11-00094.1, 2007. a
Taylor, K. E., Stouffer, R. J., and Meehl, G. A.: An overview of CMIP5 and the experiment design, B. Am. Meteorol. Soc., 93, 485–498, https://doi.org/10.1175/BAMS-D-11-00094.1, 2012. a
van Vuuren, D. P., Edmonds, J., Kainuma, M., Riahi, K., Thomson, A., Hibbard, K., Hurtt, G. C., Kram, T., Krey, V., Lamarque, J. F., Masui, T., Meinshausen, M., Nakicenovic, N., Smith, S. J., and Rose, S. K.: The representative concentration pathways: An overview, Climatic Change, 109, 5–31, https://doi.org/10.1007/s10584-011-0148-z, 2011. a
Yool, A., Popova, E. E., Coward, A. C., Bernie, D., and Anderson, T. R.: Climate change and ocean acidification impacts on lower trophic levels and the export of organic carbon to the deep ocean, Biogeosciences, 10, 5831–5854, https://doi.org/10.5194/bg-10-5831-2013, 2013. a
Zweng, M. M., Reagan, J., Antonov, J., Mishonov, A., Boyer, T., Garcia, H., Baranova, O., Johnson, D., Seidov, D., and Bidlle, M.: World Ocean Atlas 2013, Volume 2: Salinity, NOAA Atlas NESDIS, 2, 227–237, 2013. a
- Abstract
- Introduction
- The BGC-val toolkit design features
- Evaluation process
- Code structure and functionality
- Applying BGC-val to CMIP5 RCP8.5
- Conclusions
- Code availability
- Appendix A: Installing and running BGC-val
- Author contributions
- Competing interests
- Disclaimer
- Acknowledgements
- References
- Supplement
- Abstract
- Introduction
- The BGC-val toolkit design features
- Evaluation process
- Code structure and functionality
- Applying BGC-val to CMIP5 RCP8.5
- Conclusions
- Code availability
- Appendix A: Installing and running BGC-val
- Author contributions
- Competing interests
- Disclaimer
- Acknowledgements
- References
- Supplement