the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
A machine learning approach targeting parameter estimation for plant functional type coexistence modeling using ELM-FATES (v2.0)
Yilin Fang
Zhonghua Zheng
Mingjie Shi
Marcos Longo
Charles D. Koven
Jennifer A. Holm
Rosie A. Fisher
Nate G. McDowell
Jeffrey Chambers
L. Ruby Leung
Tropical forest dynamics play a crucial role in the global carbon, water, and energy cycles. However, realistically simulating the dynamics of competition and coexistence between different plant functional types (PFTs) in tropical forests remains a significant challenge. This study aims to improve the modeling of PFT coexistence in the Functionally Assembled Terrestrial Ecosystem Simulator (FATES), a vegetation demography model implemented in the Energy Exascale Earth System Model (E3SM) land model (ELM), ELM-FATES. Specifically, we explore (1) whether plant trait relationships established from field measurements can constrain ELM-FATES simulations and (2) whether machine learning (ML)-based surrogate models can emulate the complex ELM-FATES model and optimize parameter selections to improve PFT coexistence modeling. We conducted three ensembles of ELM-FATES experiments at a tropical forest site near Manaus, Brazil. By comparing the ensemble experiments without (Exp-CTR) and with (Exp-OBS) consideration of observed trait relationships, we found that accounting for these relationships slightly improves the simulations of water, energy, and carbon variables when compared to observations but degrades the simulation of PFT coexistence. Using ML-based surrogate models trained on Exp-CTR, we optimized the trait parameters in ELM-FATES and conducted another ensemble of experiments (Exp-ML) with these optimized parameters. The proportion of PFT coexistence experiments significantly increased from 21 % in Exp-CTR to 73 % in Exp-ML. After filtering the experiments that allow for PFT coexistence to agree with observations (within 15 % tolerance), 33 % of the Exp-ML experiments were retained, which is a significant improvement compared to the 1.4 % in Exp-CTR. Exp-ML also accurately reproduces the annual means and seasonal variations in water, energy, and carbon fluxes and the field inventory of aboveground biomass. This study represents a reproducible method that utilizes machine learning to identify parameter values that improve model fidelity against observations and PFT coexistence in vegetation demography models for diverse ecosystems. Our study also suggests the need for new mechanisms to enhance the robust simulation of coexisting plants in ELM-FATES and has significant implications for modeling the response and feedbacks of ecosystem dynamics to climate change.
- Article
(7691 KB) - Full-text XML
-
Supplement
(12876 KB) - BibTeX
- EndNote
Tropical ecosystems feature the highest biodiversity on Earth, maintaining more than 75 % of all known species (Mora et al., 2011; Mitchard, 2018). The dynamics of tropical forests are closely related to the regional and global carbon, energy, and water cycles (Bonan, 2008; Piao et al., 2020). Vegetation is expected to face more water stress from vapor pressure deficit increase and soil moisture reduction with global warming (McDowell et al., 2020). Tree mortality rates are accelerating in some tropical regions due to rising atmospheric water stress (Bauman et al., 2022; Hubau et al., 2020; Zuleta et al., 2017). Tropical forests currently make an approximately neutral contribution to the global carbon cycle as a result of a large land use source balanced by sinks in recovering and undisturbed forests, but they may become a carbon source in the future under the threat of climate change and human-induced disturbance (Mitchard, 2018; Gatti et al., 2021). Therefore, both understanding and modeling tropical forest dynamics and related feedbacks have crucial implications for projecting future changes in the global climate system.
Dynamic global vegetation models (DGVMs) are the primary tools to simulate terrestrial ecosystem dynamics of plant functional type distribution, ecosystem composition and functioning, and ecosystem response to and recovery from disturbance (e.g., fire and wind damage) (Longo et al., 2019; Fisher et al., 2018; Foley et al., 1996; Sitch et al., 2003; Cao and Woodward, 1998; Berzaghi et al., 2019; McMahon et al., 2011). Conventional DGVMs represent plant communities using an area-averaged representation of plant functional types (PFTs) in each grid cell. Their relatively simple structures have the advantage of high computational efficiency for use in Earth system models (Fisher et al., 2018; Snell et al., 2014). However, these models do not capture many demographic processes. For example, plants of each represented PFT typically have identical properties (e.g., tree size), which limits the capability of modeling ecosystem dynamics and functioning of canopy gap formation, PFT competition, and disturbance reactions (Feeley et al., 2007; Stark et al., 2012; Hurtt et al., 1998; Moorcroft, 2003; Brister et al., 2020). To address these limitations, researchers have developed a new of generation DGVMs called vegetation demographic models (VDMs), commonly including individual-based models and cohort-based models (Fisher et al., 2018). The individual-based models, also known as forest gap models, explicitly represent vegetation as individual plants and simulate their birth, growth, and death (Fyllas et al., 2014; Christoffersen et al., 2016; Sato et al., 2007; Jonard et al., 2020; Maréchaux and Chave, 2017). These models incorporate the stochasticity and heterogeneity of the plant light environment mechanistically and thereby can typically represent PFT competitive exclusion, succession, and coexistence. However, explicit simulations of individual plants with stochastic processes suffer a substantial computational penalty and limit applicability over large or global scales (Fisher et al., 2018). To capture sufficient ecosystem dynamics and maintain relatively high computational efficiency, “cohort-based” models have been proposed (Haverd et al., 2013; Medvigy et al., 2009; Ma et al., 2022; Moorcroft et al., 2001; Weng et al., 2015; Longo et al., 2019; Martín Belda et al., 2022). In cohort-based approaches, individual plants are grouped together as “cohorts” based on their similar properties, including size, age, and PFT (Fisher et al., 2018). Many cohort-based models have been developed and widely used across regional to global scales. Examples of cohort-based models include the Ecosystem Demography model (ED) (Moorcroft et al., 2001), the Functionally Assembled Terrestrial Ecosystem Simulator (FATES) (Fisher et al., 2018, 2015), and the Geophysical Fluid Dynamics Laboratory (GFDL) Land Model 3 with the Perfect Plasticity Approximation (LM3-PPA) (Weng et al., 2015). In this study, we employ the FATES model, a widely used tool for modeling ecosystem dynamics in multiple ecosystems, including tropical (Holm et al., 2020; Koven et al., 2020; Chitra-Tarak et al., 2021; Cheng et al., 2021), boreal (Lambert et al., 2022), and mixed-conifer forests (Buotte et al., 2021) and forest disturbance (Huang et al., 2020).
Despite ongoing applications, robust simulations of competition and coexistence in cohort-based VDMs remain a major challenge. In niche-based coexistence theory, coexisting species require both convergence in strategy to adapt to the surrounding environment (“environmental filtering”) and divergence in strategy to ensure differentiation in resource requirements (“niche partitioning”) (Kraft et al., 2008; Adler et al., 2013). These same constraints apply to coexisting PFTs as modeled by VDMs. Thus, on the one hand, VDMs need to include mechanisms that capture critical niche dimensions (e.g., spatial and temporal variation in light, water, and nutrients). For example, the multilayer canopy structure in FATES provides vertical light resource differentiation. Another essential aspect is to assign reasonable plant functional traits (i.e., the parameters that define a given plant functional type) to satisfy environmental filtering, ensure niche partitioning, and consequently preserve PFT coexistence. Considering the relatively high computational cost of VDMs and the host land surface models, it is not feasible to directly apply global optimization methods such as shuffled complex evolution (Duan et al., 1992) to calibrate trait-related parameters because this could be a time-consuming and computationally intensive process (Rouholahnejad et al., 2012). Therefore, most previous studies use the filtered ensemble approach to select trait-related parameters involving several steps: (1) generating a parameter ensemble based on reference trait ranges or correlations, (2) conducting ensemble model simulations, and (3) filtering the parameter ensemble by coexistence and other criteria (e.g., observation constraints). For example, Huang et al. (2020) applied FATES implemented in the Community Land Model (CLM; herein CLM-FATES) with two tropical PFTs to study forest dynamics at tropical sites. They performed 70 one-at-a-time experiments before obtaining one reasonable parameter set. Buottte et al. (2021) used CLM-FATES to simulate forest dynamics of pine and incense cedar over the Sierra Nevada of California, and their two stages of experiments (360 plus 72 runs) only yielded four sets of parameters that met the given criteria. The filtered ensemble approach has low efficiency, which hinders VDMs' application to modeling ecosystem dynamics under the changing climate. In addition, trait relationships derived from field measurements are often used to infer parameter selections when simulating coexistence. For example, Longo et al. (2020) used multiple trait relationships derived from various datasets to guide parameter selection for different PFTs in the ED-2.2 model simulations. However, whether the observed trait relationships can efficiently improve PFT coexistence simulation in current VDMs is still unclear. Earlier studies using FATES have also highlighted the importance of reproductive feedbacks in maintaining or prohibiting coexistence (Fisher et al., 2010; Maréchaux and Chave, 2017). Fundamentally, if PFTs have highly contrasting reproductive output, the model tends towards competitive exclusion, and thus discerning areas with at least approximately equal fitness is necessary. While representing a large number of plant functional types may improve the likelihood of coexistence (Koven et al., 2020), this comes at a considerable computational expense.
Machine learning (ML) has facilitated Earth science studies (Shen, 2018; Nearing et al., 2021; Zhu et al., 2022; Pal et al., 2019; Jung et al., 2019), possibly providing a promising approach to improve PFT coexistence modeling in VDMs. ML algorithms have been broadly and successfully employed in recent decades. They can be used as standalone models to predict variables of interest or integrated with process-based models to improve simulations (Xu and Liang, 2021; He et al., 2022; Peatier et al., 2022). Among these applications, ML has shown advantages as a surrogate model for parameter optimization and sensitivity quantification, including its effectiveness and easy application, its ability to implicitly deal with complex nonlinear correlations and high dimensional data, and handle interactions between variables (Sit et al., 2020; Antoniadis et al., 2020; Tsai et al., 2021). One promising approach is to construct ML-based surrogate models using data from initial model simulations to emulate the relationship between inputs (i.e., model parameters) and model outputs (Wang et al., 2014). The computationally inexpensive surrogate model can then be efficiently used for parameter optimization and sensitivity analysis. For example, Dagon et al. (2020) implemented artificial neural networks to emulate the satellite leaf area constrained version of CLM5 (Lawrence et al., 2019) and estimated optimal parameters to improve the global simulation of gross primary production and latent heat flux. Sawada (2020) developed an ML surrogate model to optimize the land surface model parameters and improve soil moisture and vegetation dynamics simulations. Watson-Parris et al. (2021) built a general tool to efficiently emulate Earth system models for uncertainty quantification and model calibration. Although employing ML-based surrogate models to optimize the trait parameters and hence improve the vegetation dynamics modeling in VDMs is promising, this area of research remains under-explored.
This study aims to improve PFT coexistence modeling in VDMs. The cohort-based FATES implemented in the Energy Exascale Earth System Model (E3SM) land model (ELM; Golaz et al., 2019), i.e., ELM-FATES, is taken as our test bed. The ELM land model simulates surface energy fluxes, soil and canopy biophysics, hydrology, and soil biogeochemistry, whereas FATES simulates live vegetation processes, litter dynamics, and fire. We first examine whether trait relationships constructed from field measurements can help improve ELM-FATES simulations. Second, we explore whether ML-based surrogate models can help optimize key trait parameters in ELM-FATES to improve the simulation of PFTs coexistence. Our model experiments are conducted for a tropical rainforest site located in Manaus, Brazil. This paper is organized as follows. Section 2 describes ELM-FATES, summarizes the key functional trait-related parameters, introduces the machine learning algorithms, and explains the overall experimental design. Results are presented in Sect. 3, followed by discussions and conclusions in Sects. 4 and 5, respectively.
2.1 Study site and data
Our study site is located at kilometer 34 (K34) of the ZF2 road, Manaus, Brazil (latitude: −2.6091∘ S; longitude: −60.2093∘ W). The K34 site is an old-growth primary forest with minimal human disturbances (Holm et al., 2020). The annual precipitation is about 2252 mm, and the mean temperature is about 26.68 ∘C (https://ameriflux.lbl.gov/sites/siteinfo/BR-Ma2, last access: 6 June 2022). The wet season is from November to May, and the dry season is from June to October (Fang et al., 2017). Hourly meteorological forcing (i.e., precipitation, air temperature, relative humidity, wind speed, surface pressure) at the K34 eddy covariance flux tower from 2002 to 2005 was obtained from the LBA-ECO CD-32 Flux Tower Network Data Compilation (Restrepo-Coupe et al., 2021). Observational reference datasets obtained from Holm et al. (2020) include gross primary production (GPP), evapotranspiration (ET), sensible heat flux (SH), Bowen ratio (BW, the ratio between sensible heat and latent heat), and inventory data-based aboveground biomass (AGB). The GPP, ET, SH, and BW observations are monthly climatological averages from 2000 to 2008 (Table S1). The AGB at this site is about 303±2.3 Mg ha−1. These observational data were used to evaluate the ELM-FATES simulations and constrain the ML surrogate models.
2.2 ELM-FATES and parameters
ELM-FATES is used as the model test bed. ELM is the land model of E3SM, which is the host land model of FATES (Golaz et al., 2019; Leung et al., 2020; Holm et al., 2020). FATES is a size- and age-structured vegetation model developed from the Community Land Model with Ecosystem Demography (CLM-ED) (Fisher et al., 2015; Koven et al., 2020). FATES includes two key structural components: ecosystem demography (ED; Moorcroft et al., 2001) and a modified version of perfect plasticity approximation (PPA, Purves et al., 2008). FATES discretizes the simulated landscape into spatially implicit patches representing different disturbance histories of the ecosystem since the last disturbance. Within each patch, the hypothetical population of plants is grouped into cohorts: a cohort consists of a population density of trees with similar size and the same plant functional type. Cohorts are organized, via the PPA concept, into canopy layers, and compete for light based on their canopy vertical positions (e.g., canopy layer vs. understory layer). The understory layer is formed when the canopy area becomes greater than the total ground area, and some fraction of each cohort is “demoted” to the understory as a function of its height. The number of patches and cohorts varies depending on processes, including recruitment, growth, mortality, competition, and disturbance. The modified PPA probabilistically splits cohorts into discrete canopy and understory layers based on a function of their height (Strigul et al., 2008; Fisher et al., 2010). A detailed description of the FATES model can be found in its technical note (Zenodo, https://doi.org/10.5281/zenodo.3517272; FATES Development Team, 2019).
In this study, we configured two PFTs in ELM-FATES, i.e., early successional and late successional broadleaf evergreen tropical trees, which can represent a primary axis of variability in tropical forests (Huang et al., 2020; Reich, 2014; Díaz et al., 2016). There are tradeoffs between the plant traits of these two PFTs. Compared with the late successional PFT, the early successional PFT is more light demanding and fast growing but with lower woody density, shorter leaf and root lifespans, and higher background mortality. To represent the drought impacts on forest dynamics, the early successional PFT is further assumed to be less drought resistant with shallower rooting depth and is hence more easily affected by drought conditions (Oliveira et al., 2021). The corresponding tradeoffs and parameters between these two PFTs are shown in Fig. 1 and Table 1.

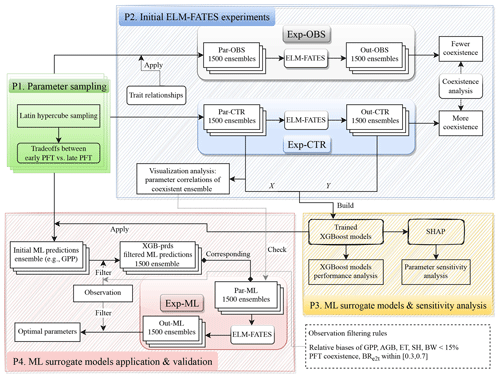
Figure 1Schematic representation of tradeoffs between early and late successional PFTs. Dark red denotes a higher parameter value. The tradeoffs of the top five traits are used to constrain the parameter sampling.
Table 1Summary of ELM-FATES trait parameters for two PFTs.
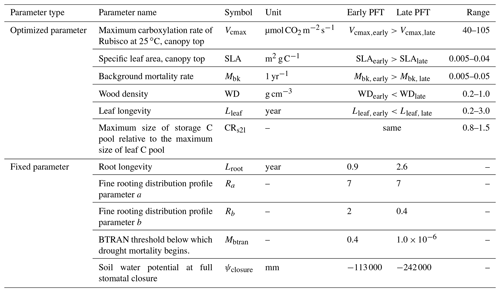
Parameter references (Huang et al., 2020; Koven et al., 2020; Longo et al.,
2020; Holm et al., 2020; Cheng et al., 2021; Domingues et al., 2005;
Chitra-Tarak et al., 2021; Buotte et al., 2021).
Ra and Rb are parameters that determine the rooting depth and
vertical distribution of fine roots.
BTRAN is the plant water stress factor. BTRAN , where 0
represents full water stress and 1 represents no water stress.
2.3 Machine learning algorithm
We built ML-based surrogate models to emulate ELM-FATES simulations. To represent the relationships between ELM-FATES parameters and simulations (e.g., AGB), we used eXtreme Gradient Boosting (XGBoost; Chen and Guestrin, 2016), a decision-tree-based ensemble machine learning algorithm. Ensemble learning techniques combine the predictions of multiple independent base models (e.g., decision trees) to produce more accurate predictions, with popular algorithms such as Random Forest (Breiman, 2001) and XGBoost. While Random Forest builds an ensemble of parallel trees using bagging and produces the final prediction by averaging the outputs of all individual trees, XGBoost sequentially trains a set of decision trees using boosting (Friedman, 2001), where each successive tree corrects the mistakes of its predecessors, and the final prediction is obtained by combining the predictions of all trees using a weighted sum. XGBoost not only handles complex nonlinear interactions and collinearity between different features but also provides a parallel implementation that effectively solves a range of data science problems. It has been successfully applied in a variety of fields within Earth and Environmental Sciences, such as urban temperature emulation (Zheng et al., 2021c), wildfire-burned area (Wang et al., 2021), emissions prediction (Wang et al., 2022), flash flood risk assessment (Ma et al., 2022), and aerosol property estimation (Zheng et al., 2021a, b).
2.4 Experimental design
The experimental design flowchart is shown in Fig. 2. Overall, we generated three ensembles of parameter values, i.e., Par-CTR, Par-OBS, and Par-ML, and conducted three ensembles of corresponding ELM-FATES experiments, i.e., Exp-CTR, Exp-OBS, and Exp-ML. Exp-CTR is the control experiment without being constrained by the observed trait relationships. Exp-OBS considered the constraint of the observed trait relationships. Par-ML was generated by machine learning surrogate models, which were trained based on Exp-CTR, and then used to conduct Exp-ML. The detailed experiment procedures are described below.
2.4.1 Procedure 1: parameter sampling
The procedure “P1” in Fig. 2 is used to generate an ensemble of parameter values for each experiment ensemble, i.e., Exp-CTR, Exp-OBS, and Exp-ML. First, a number of initial parameter sets (e.g., 5000 sets) were generated using Latin hypercube sampling (LHS; Mckay et al., 2000). Second, the initial parameter sets were filtered by the trait tradeoffs between early and late successional PFTs (Fig. 1). We repeatedly increased the number of initial parameter sets in the first step until 1500 parameter sets were obtained in the second step. Each ELM-FATES experiment starts from bare ground and runs for 350 years to reach an equilibrium state by cycling the meteorological forcing during 2002–2005, and the last 4 years of the simulations were analyzed.
2.4.2 Procedure 2: initial ELM-FATES experiments of Exp-CTR and Exp-OBS
To test whether plant trait relationships established from field measurements can improve the ELM-FATES simulations, we derived three trait relationships based on the tropical studies of Koven et al. (2020) and Longo et al. (2020). Using the digitized data from Fig. 3 in Koven et al. (2020), the background mortality Mbk (see Table 1 for parameter definitions) can be empirically computed from the maximum carboxylation rate Vcmax,
Based on the equations in Fig. S18 of Longo et al. (2020), the leaf longevity (Lleaf) and wood density (WD) can be calculated via the specific leaf area (SLA),
These trait relationships were used to generate parameters for Par-OBS.
Two initial sets of experiment ensembles, i.e., Exp-CTR and Exp-OBS (procedure “P2” in Fig. 2), were conducted based on Par-CTR and Par-OBS, respectively. For Par-CTR, 1500 parameter sets were generated from the procedure P1 based on the entire set of 11 parameters, i.e., Vcmax,early, Vcmax,late, SLAearly, SLAlate, Mbk, early, Mbk, late, WDearly, WDlate, Lleaf, early, Lleaf, late, and CRs2l (maximum size of storage C pool relative to the maximum size of leaf C pool, Table 1). For Par-OBS, 1500 parameter sets were generated from the procedure P1 but were only based on five parameters (i.e., Vcmax,early, Vcmax,late, SLAearly,SLAlate, and CRs2l). The other six parameters (Mbk, early, Mbk, late, WDearly, WDlate, Lleaf, early, and Lleaf, late) in Par-OBS were calculated based on the trait relationships defined by Eqs. (1)–(3). Therefore, compared to Par-CTR, the parameters in Par-OBS are constrained by the observed trait relationships. The distributions of these two parameter sets are shown in Fig. S1. Vcmax, SLA, and CRs2l have similar distributions between Par-CTR and Par-OBS. Compared with Par-CTR, Par-OBS has a narrower distribution of Mbk but broader distributions of WD and Lleaf.
Exp-CTR and Exp-OBS each include 1500 total 350-year ELM-FATES simulations. We averaged the last 4 years of these simulations for analysis, i.e., simulation outputs: Out-CTR and Out-OBS, respectively. To quantify the PFT coexistence, we computed the biomass ratio between early successional PFT and the total biomass, denoted as BRe2t. For brevity, we denote the ELM-FATES experiments with as “coexistence”, as “late”, and as “early”. We calculated BRe2t based on Out-CTR and Out-OBS and then computed the fraction of coexistence experiments in each ensemble. As we will show in Sect. 3.1, when considering the observed trait relationships, Exp-OBS has a lower fraction of coexistence experiments. Therefore, only Exp-CTR was used for further ML-related analysis. We also performed some analysis of Exp-CTR to explore whether the parameters of the coexistence experiments have correlations with each other (Sect. 3.2).
2.4.3 Procedure 3: ML surrogate models and sensitivity analysis
Based on Exp-CTR, we trained XGBoost models to emulate the ELM-FATES model behavior and analyzed the parameter sensitivity (procedure “P3” in Fig. 2). A total of 16 variables were used as XGBoost model features, including 11 parameters in Par-CTR and 5 parameter differences between early and late successional PFTs. The corresponding ELM-FATES annual average outputs were used as XGBoost model targets. Specifically, six models were built, i.e., XGB_ET, XGB_SH, XGB_BW, XGB_GPP, XGB_AGB, and XGB_BR for predicting ET, SH, BW, GPP, AGB, and BRe2t, respectively. The ML models were trained, tested, and subsequently utilized to perform the parameter sensitivity analysis, as described in Sect. 2.5.
2.4.4 Procedure 4: ML surrogate models application and validation
The trained XGBoost models were then used to help select ELM-FATES parameters (procedure “P4” in Fig. 2). First, initial parameter sets were generated from procedure P1 based on the entire set of 11 parameters (Table 1, identical to the parameter set used for the generation of Par-CTR). Second, these parameter sets and parameter differences were sent to six XGBoost surrogate models to predict ET, SH, BW, GPP, AGB, and BRe2t. Third, the predictions were further filtered by two criteria: (1) compared to observations, the relative biases of the predicted ET, SH, BW, GPP, and AGB should be less than 15 %, and (2) the XGBoost model-predicted BRe2t should be within [0.3, 0.7], which corresponds to the range where the XGB-BR model exhibited relatively better performance (Fig. 5). We repeated these three steps until we obtained 1500 sets of XGBoost model predictions that matched the criteria. Finally, we obtained 1500 sets of XGBoost model predictions and their corresponding 1500 sets of parameters (Par-ML). We also checked whether the selected Par-ML could match the empirical relationships derived from the empirical analysis in procedure P2 (see Sect. 3.2 and 3.5 for details). Following this, the 1500 sets of parameters in Par-ML were sent to ELM-FATES to conduct 350-year runs (i.e., Exp-ML). The last 4 years of the simulations were averaged (i.e., Out-ML) for further analysis. We then filtered Out-ML based on a relative bias of 15 % or less compared to observations and PFT coexistence to identify the optimal experiments and corresponding parameters.
2.5 ML model development and SHAP analysis
The process of building each of the six ML surrogate models is described. Taking BRe2tas an example, the 1500 pairs of 16 features and the corresponding simulated BRe2t were randomly split into two groups, with 90 % used for training and the remaining 10 % used for testing. Given that the coexistence experiments only account for 20.6 % in the simulations of Exp-CTR (Sect. 3.1), we used 90 % of the data for training to ensure sufficient coexisting samples were included in the training process. Optimizing the hyperparameters of the XGBoost model is crucial for its performance. To achieve this, we employed the Bayesian optimization method during the training process (Snoek et al., 2012). In addition, to avoid overfitting during hyperparameter optimization, we utilized a 5-fold cross-validation method (Feigl et al., 2021). The mean squared error was used as the objective function to achieve the optimal hyperparameters. The root-mean-squared error (RMSE) and R-squared value (R2) are used to quantify the overall model performance for the training and testing data prediction.
Based on the trained XGBoost models, we subsequently employed a game theory approach called SHapley Additive exPlanations (SHAP; Lundberg and Lee, 2017; Lundberg et al., 2018, 2020) to gain insights into the parameter sensitivity of ELM-FATES. SHAP assumes that features (predictive variables) interact and collaborate in a prediction game, with each feature receiving a payout for its contributions. This approach provides a unified measure of feature importance to explain both individual samples and the entire dataset, which is distinct from intrinsic feature importance methods such as the feature importance in XGBoost (Lundberg and Lee, 2017). This approach has been widely used in various fields, including interpreting a digital soil mapping model (Padarian et al., 2020) and identifying the critical drivers of wildfires (Wang et al., 2021). In this study, we performed SHAP analysis for each XGBoost model and used the SHAP values as a proxy to quantify the relative importance of ELM-FATES parameters.
3.1 Comparison between Exp-CTR and Exp-OBS
Constraining the input traits using the observed trait relationships yields slightly better ELM-FATES simulations of water, energy, and carbon variables (Fig. 3a–e). The distributions of the relative biases of ET, SH, BW, and GPP have similar ranges between the two sets of experiments (Fig. 3a–d). Compared with Exp-CTR, the 50th percentiles of relative biases of ET, SH, BW, and GPP for Exp-OBS (with constrained traits) are closer to zero, indicating Exp-OBS is slightly better than Exp-CTR. The distribution of simulated AGB for Exp-OBS is much narrower than Exp-CTR (Fig. 3e), which could be due to the narrower distribution of Mbk (Fig. S1).
Exp-CTR has a much higher fraction of coexisting PFT simulations than Exp-OBS (Fig. 3f and Table S2). Overall, 70.6 % of experiments in Exp-CTR and 94.5 % of experiments in EXP-OBS have simulated BRe2t that is greater than 0.9. This indicates that both Par-CTR and especially Par-OBS favor the early successional PFT. As for the coexisting experiments with , Exp-CTR has about 5 times more coexisting experiments (20.6 %) than Exp-OBS (4.1 %). Further filtering the coexisting cases by observations (Table S1), only 21 experiments remain in Exp-CTR, and only 6 experiments remain in Exp-OBS (Table S2). Even though Exp-OBS considered the observed trait relationships, it has fewer coexisting cases within the reasonable observation ranges than Exp-CTR. Therefore, Exp-OBS is not used in our remaining analysis.
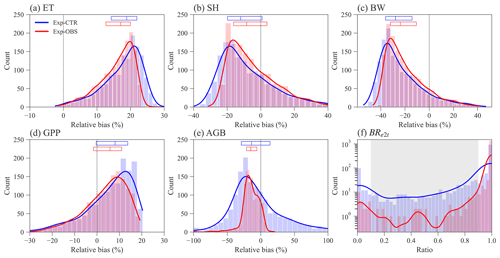
Figure 3Distribution of ELM-FATES simulations for Exp-CTR and Exp-OBS. The y axis in (f) is logarithmic. Relative bias is equal to (%). In (a)–(e), the top horizontal bars with three vertical lines denote the relative bias at the 25th, 50th, and 75th percentiles, respectively. The grey-shaded area in (f) represents the coexistence biomass ratio between 0.1 and 0.9.
3.2 Parameter analysis of Exp-CTR
We also tested whether simple parameter correlations can be constructed to guide the simulation of PFTs coexistence. No simple parameter correlations can be built to distinguish the coexisting cases from the early and late cases in Exp-CTR (Figs. 4, S2, and S3). Most parameter (or parameter difference) spaces show large overlaps between early, late, and coexisting cases (Figs. S2 and S3). Notably, we empirically built three linear equations based on the boundaries in the parameter spaces for the coexisting cases (Fig. 4). Coexisting cases are primarily located in spaces with (Fig. 4a and d), (Fig. 4b and e), and (Fig. 4c and f), where , and SLAdiff and WDdiff are defined likewise. Within these constrained parameter spaces, the percentage of coexisting cases increases from the original 20.6 % (i.e., 309 out of 1500) to 32.6 % (i.e., 304 out of 932). Therefore, these empirical correlations could help guide ELM-FATES parameter selection for coexisting PFTs. On the other hand, a dominant proportion (i.e., 67.4 % (100 %–32.6 %)) of experiments are still either early or late cases within the constrained parameter spaces and cannot robustly predict PFT coexistence. Moreover, despite further considering the observational constraints (black scatters in Fig. 4; Table S2), the 21 experiments (2.3 %, 21 out of 932) are still sparsely distributed in the parameter space of the coexisting cases, so no simple correlations can be developed based on these simulations. Therefore, simple empirically built relationships between plant traits provide limited benefit to guiding ELM-FATES parameter selection for modeling PFTs coexistence while matching the observations. This finding provides additional motivation for the ML-based approaches.
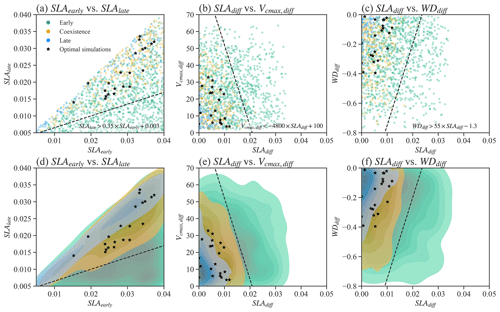
Figure 4Relationships between selected parameters of Par-CTR. These parameters are presented in three groups, i.e., green color for the late cases with , orange color for the coexisting cases with , and blue color for the early cases with . Black stars represent coexistence cases further filtered by observational constraints. Panels (d)–(f) are the corresponding kernel density estimate plots of the scatter plots shown in (a)–(c). . SLAdiff and WDdiff are defined likewise.
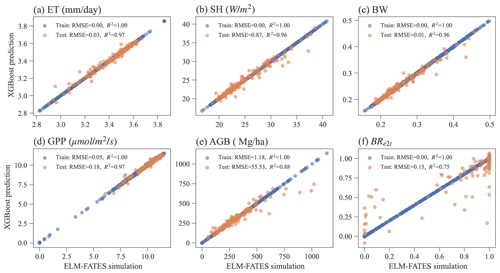
3.3 XGBoost model performance
Overall, the XGBoost surrogate models show good performance in predicting ELM-FATES simulations (Fig. 5). Based on Exp-CTR (i.e., Par-CTR and Out-CTR), six XGBoost models were trained. In training, the RMSEs for the six models are zero or nearly zero, and R2s are close to one. In the testing, four XGBoost models (i.e., XGB_ET, XGB_SH, XGB_BW, XGB_GPP) still show good performance with small RMSE and large R2 (>0.95). XGB_AGB shows a little degradation with R2 of 0.88. The performance of XGB_BR also shows degradation, with R2 decreasing from 1.0 in training to 0.75 in testing. XGB_BR cannot predict the ELM-FATES-simulated BRe2t of 0 or 1 well when only one PFT survives. This indicates that PFT competition processes in ELM-FATES, which determine BRe2t and AGB, are highly nonlinear and difficult to emulate even using a state-of-the-art machine learning algorithm.
3.4 SHAP parameter importance analysis
Figure 6 shows the feature importance, including parameters and parameter differences, for different XGBoost models. Features (on the y axis) with a higher mean absolute SHAP value (on the x axis) denote a larger contribution to the XGBoost model prediction. The number of most important features is different for predicting ET, SH, BW, and GPP compared to predicting AGB and BRe2t.
For the XGBoost models that predict ET, SH, BW, and GPP, the top three features have the largest SHAP values compared to the rest (Fig. 6a–d). Notably, these top three features are the same and correspond to the early successional PFT, i.e., Vcmax,early, SLAearly, and Lleaf, early. Most ELM-FATES experiments in Exp-CTR used as the training samples for the XGBoost models are early cases. Therefore, the parameters of early successional PFT have dominant contributions in the XGBoost model predictions of overall grid-level fluxes. These three parameters are positively correlated with ET and GPP and negatively correlated with SH and BW (red vs. blue bars in Fig. 6a–d; see Fig. S4 for more details), reflecting the fundamental carbon metabolism of the typically dominant early successional plant.
For the XGBoost surrogate models of AGB and BRe2t, more than eight features have large SHAP values (Fig. 6e and f). Both early and late successional PFT parameters contribute to predicting the two variables. Compared with the predictions of ET, SH, BW, and GPP with only three major features, predicting AGB and BRe2t is relatively complex. This is because AGB and particularly BRe2t are closely related to the PFT competition process in which both the early and late PFT traits are crucial. Especially for BRe2t, the most important features are the parameter difference between the early and late successional PFTs. For example, SLAdiff is positively correlated to BRe2t. Therefore, to have coexisting PFTs with , the SLA of two PFTs should be neither too large nor too small.
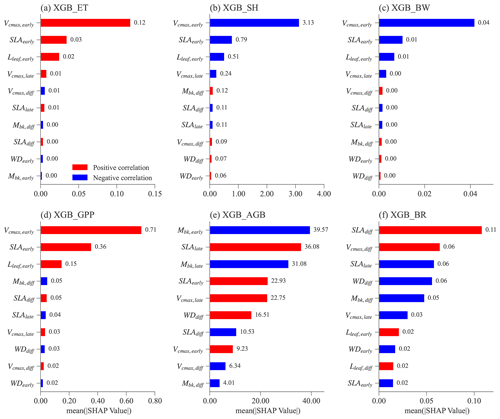
Figure 6Mean absolute SHAP values for different XGBoost surrogate models for the top 10 most important features. Absolute SHAP values are sorted in decreasing order from top to bottom. For each feature (y axis) in each XGBoost model, the Spearman correlation coefficient is calculated between the feature values and the corresponding SHAP values (Fig. S4). The red color means that a given feature is positively correlated with the predicting variable, whereas blue denotes a negative correlation.
3.5 XGBoost model parameter selection
Using the XGBoost surrogate models, the Par-ML was selected, including 1500 sets of parameters and the corresponding parameter differences between the early and late successional PFTs (Sect. 2.4, procedure P4 in Fig. 2). We examined whether Par-ML matches the empirical relationships shown in Fig. 4 (Sect. 3.2), i.e., , , and . In total, 99.1 % (1486 out of 1500) of parameter sets are consistent with the empirical relationships, indicating the XGBoost models implicitly learned these simple relationships.
The parameter distributions of Par-ML show different patterns from the early and late parameters of Par-CTR (green vs. blue regions in Fig. 7), but there are large overlaps between the coexistence parameters of Par-CTR and Par-ML (orange vs. green regions, e.g., the third column in Fig. 7). This indicates that the XGBoost surrogate models learned to select parameters around the parameters' space of the coexisting cases. Par-ML also tends to have a smaller parameter difference between the early and late successional PFTs in terms of SLAdiff and Vcmax, diff. However, Par-ML also shows different patterns from the coexisting parameters of Par-CTR, probably because the XGBoost-selected parameters were also constrained by multiple observations and implicitly considered parameter tradeoffs. For example, the Vcmax,early and Vcmax, late of Par-ML are located in narrower ranges than the coexisting parameters of Par-CTR (first two columns in Fig. 7).
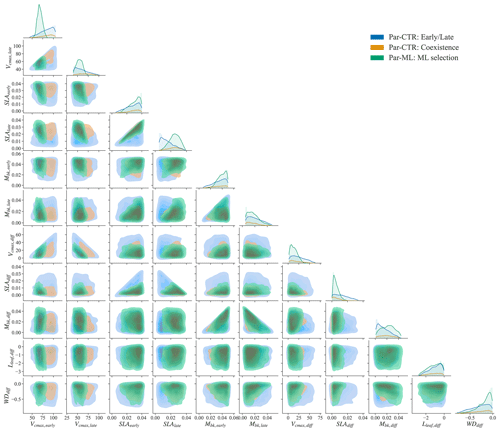
Figure 7Comparison of parameter or parameter difference in Par-CTR vs. Par-ML for 11 features. The diagonal plots represent each parameter's distribution, and the rest of the subplots are kernel density estimate plots. There are three groups, i.e., blue for the early and late cases of Par-CTR, orange for the coexisting cases of Par-CTR, and green for Par-ML selected by XGBoost models.
3.6 Validation of ML selected parameters
ELM-FATES simulations of Exp-ML based on the ensemble parameters of Par-ML selected by the XGBoost surrogate models can better capture the observations and have more coexisting cases than Exp-CTR (Fig. 8). The median values of simulated variables for Exp-ML are closer to observations with relative biases closer to zero than Exp-CTR (Fig. 8a, blue vs. green boxes). The Exp-ML-simulated variables also have more concentrated distributions than Exp-CTR. Compared to the skewed distribution of BRe2t in Exp-CTR with a large proportion of early cases, Exp-ML has a more normally distributed BRe2t (Fig. 8b). Specifically, Exp-ML has about 3.6 times more coexisting cases than Exp-CTR, i.e., 73.1 % (1097 out of 1500) in Exp-ML vs. 20.6 % (309 out of 1500) in Exp-CTR (Table S3). After being further constrained by observations (Table S3), one-third of the experiments (i.e., 495 out of 1500) in Exp-ML remain, and this ratio is 23.6 times more than 1.4 % (21 out of 1500) in Exp-CTR.
The XGBoost surrogate model-predicted variables also match well with those simulated using ELM-FATES in Exp-ML (Fig. 8, orange vs. green boxes), indicating the overall reasonable accuracy for the XGBoost model predictions. Compared to the ELM-FATES results using Par-ML, the XGBoost models show better performance for ET, SH, BW, and GPP but relatively degraded performance for AGB and BRe2t (Fig. S5). It is consistent with the performance of the XGBoost models' training and testing results (in Sect. 3.3).

Figure 8Comparison between the ELM-FATES simulations for Exp-CTR and Exp-ML. (a) Relative bias for simulated ET, SH, BW, GPP, and AGB. (b) Simulated BRe2t. ML prediction represents the selected XGBoost model predictions after filtering with observation and biomass ratio (i.e., the XGB_prds, procedure P4 in Fig. 2).
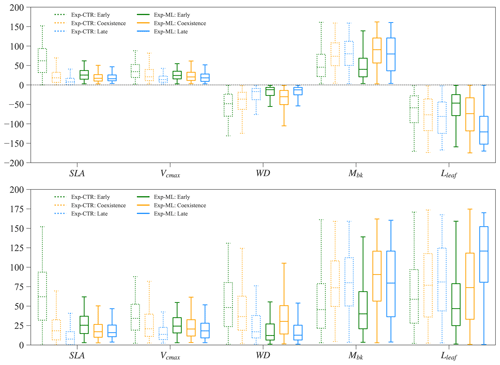
3.7 Parameter tradeoff for coexistence experiments
Parameters of the early and late successional PFTs show tradeoffs for the coexisting experiments. Large relative differences in SLA, Vcmax, and WD (more negative) favor the early successional PFT, while large relative differences in Mbk and Lleaf favor the late successional PFT. Therefore, in Exp-CTR, compared to the early and late cases, the coexisting cases have intermediate relative differences in SLA, Vcmax, WD, Mbk, and Lleaf (dashed boxes in Fig. 9). The coexisting cases in Exp-ML have similar patterns with intermediate relative differences in SLA, Vcmax and Lleaf compared to the early and late cases (solid boxes in Fig. 9). However, Mbk and especially WD show the largest relative difference for the coexisting cases compared to the early and late cases in Exp-ML. These two parameters still show a tradeoff in determining coexisting PFTs because larger WD favors the early PFT, whereas larger Mbk favors the late PFT.
In Exp-ML, the parameter spaces of the coexisting cases show large overlaps with the early and late cases (Fig. S6). There are no simple correlations between these parameters to distinguish the coexisting cases from the early and late cases (also see Sect. 3.2). Although WDdiff of the coexisting cases still overlap with the early and late cases, when WDdiff is less than roughly −0.4 g cm−3, only coexisting cases exist (Fig. S6). Nevertheless, this rule (i.e., ) alone cannot ensure PFT coexistence (see Fig. 7).
3.8 Seasonal variation comparison
Figure 10 shows the seasonal variations in ET, SH, BW, and GPP for observations and simulations of the finally selected 495 experiments in Exp-ML with good model performance (Table S3). Overall, the simulated ET shows a similar seasonal variation to ET observation (Fig. 10a), with relatively small ET in the wet season (November–May), high ET in the dry season (June–October), and ET peaks in August. However, compared to the observations, ELM-FATES overestimates ET, especially during the wet season. The simulated SH also shows a similar seasonal variation with the SH observation except in March. ELM-FATES overestimated SH from January to May but underestimated SH from September to December (Fig. 10b). Due to the discrepancy between simulated ET and SH, the model underestimates BW from September to December (Fig. 10c).
The simulated GPP has minor seasonal variability compared to the observed GPP. ELM-FATES overestimates GPP from June–August in the dry season but underestimates GPP over October–December. The lower GPP over June–August indicates that plants may be relatively water stressed or energy limited during these months. However, the large ET observation over the same period implies that this site is unlikely to be water limited or strongly energy limited. The ELM-FATES simulations also display little water stress year-round (Fig. S7). Therefore, there are likely elements of the seasonal cycle (e.g., phenological responses of photosynthetic capacity) that are not yet captured here. Additionally, tower estimates of GPP may also have large uncertainties.
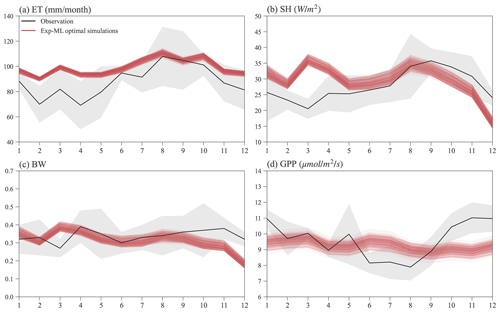
Figure 10Mean monthly observations and selected optimal ELM-FATES simulations in Exp-ML for (a) ET, (b) SH, (c) BW, and (d) GPP. Each red line represents one experiment simulation (4-year simulation average). The black curves are monthly climatologic averages from 2000 to 2008, and the grey-shaded area represents the interannual variabilities (i.e., mean ± standard deviation).
4.1 Limited guidance of observed trait relationships for PFT coexistence modeling in FATES
We found degraded PFT coexistence in ELM-FATES simulation when observed trait relationships are considered. More specifically, constrained by observed trait relationships, Exp-OBS has fewer coexisting cases than Exp-CTR, which does not consider the observed trait relationships. The observed trait relationships were derived from site measurements in the species-rich tropical ecosystem where plant coexistence commonly happens (Kraft et al., 2008), which is expected to enhance the PFT coexistence simulations. This inconsistency could be due to several possible reasons. First, ELM-FATES is a typical “trait filtering” model (Fisher et al., 2018), and the realistic simulation of PFT dynamics largely depends on the fidelity with which trait tradeoff surfaces are prescribed in the model (Scheiter et al., 2013). Implicit representation of trait tradeoff in the current ELM-FATES model may not be well balanced, which may differ from the observed trait relationships that lead to coexistence in the real world (at least for the ecosystem at our study site). In particular, there may be correlated tradeoffs that are measured (e.g., with below ground processes, Chitra-Tarak et al., 2021) but not represented in the model. A second reason could be the mismatch between different spatial scales. The observed trait relationships are derived from field measurements across tropical forests over a large region with diverse species and climate. For example, the relationship in Eq. (1) is for plant species in Panama. In contrast, ELM-FATES simulations were conducted at the K34 site scale with specific species composition. Therefore, the large-scale trait relationships may not reflect the small-scale trait relationships. Wright et al. (2005) showed that trait relationships fitted for individual sites varied considerably. Third, the observed trait relationships are based on simplified equations, which may not be able to comprehensively reflect PFT coexistence. For example, although Eq. (2) derived from Longo et al. (2020) can reflect the negative relationship between SLA and Lleaf, the R2 of this equation is about 0.49, which may not be accurate enough to represent trait relationships. Additionally, these Eqs. (1)–(3) do not consider the uncertainty in trait covariance. In Koven et al. (2020), the uncertainties between trait covariance were considered when sampling parameters for FATES experiments. Furthermore, machine learning models can also be employed to extract the relationships between plant traits, which can then be incorporated into ELM-FATES and evaluated in future studies.
4.2 Advantages of ML surrogate models on improving PFT coexistence modeling
ELM-FATES simulations driven by parameters selected using the XGBoost models essentially improved PFT coexistence and better captured observations. Compared to the initial Exp-CTR, which was used to train the XGBoost models, the proportion of coexisting PFTs in Exp-ML reaches 73.1 %, 3.6 times more than the 20.6 % in Exp-CTR. Further filtering the coexistence experiments by observations, Exp-ML still has 33.0 % of experiments left with good model performance, 23.6 times that of the 1.4 % of experiments in Exp-CTR with good performance. Our ML-based approach also outperforms the empirical correlations built in Sect. 3.2, which only yield 32.5 % of coexistence experiments, and this reduces to 2.3 % of experiments if further constrained by observation. The large proportion of optimal experiments selected by our ML approach also outperforms previous studies using direct filtering approaches. Buotte et al. (2021) conducted two stages of experiments to select optimal parameters for CLM-FATES modeling with two conifer species; only 0.3 % (1 out of 360) of the cases met the given criteria in the first stage of experiments, which increased to 5.5 % in the second stage of experiments. Huang et al. (2020) conducted CLM-FATES modeling with two tropical PFTs at the Tapajós National Forest sites; only 1 parameter set out of 70 (about 1.4 %) was selected with reasonable fractions of two PFTs and minor errors compared to observations. In addition, the parameter selection procedures of these two studies require some degree of subjective decision making and expert knowledge. On the other hand, our ML-based approach takes a more objective procedure, and little expert knowledge is required except for the initial determination of the parameter reference ranges. Importantly, we believe this approach can be repeatable as, e.g., model developments lead to changes between the parameter values and model predictions of forest structure and function and can be used to define constrained ensemble values that will allow assessment of confidence in model predictions. Even though simulating the coexistence of different plants may not be a big concern for individual-based VDMs, e.g., LPJmL-FIT (Sakschewski et al., 2015, 2016) and TROLL (Maréchaux and Chave, 2017), our approach also could be applied to the selection of key parameters that regulate vegetation dynamics in these models.
Our study also reproduced the observations satisfactorily. Holm et al. (2020) conducted the ELM-FATES simulation with only one PFT considered at the same K34 site. Our study yields better or similar performance in the magnitude of AGB and the magnitude and seasonal variation in GPP, ET, SH, and BW (Table 2 and Fig. 3 in Holm et al., 2020, vs. Figs. 8 and 10 in this study). It should also be noted that the overestimation of simulated energy fluxes (latent heat and SH) from January to May could be associated with the energy-related processes (e.g., energy partition, surface albedo) in ELM-FATES. Other potential reasons could be related to the uncertainties in atmospheric forcing and the common issue of incomplete energy budget closure at eddy covariance towers (Wilson et al., 2002; Foken, 2008; Da Rocha et al., 2009).
Compared to the predictions of GPP, ET, SH, and BW simulated by ELM-FATES, the XGBoost surrogate models show slightly degraded performance in predicting the simulated BRe2t and AGB (Figs. 5 and S5). Three parameters (Vcmax,early, SLAearly, and Lleaf, early) mainly control the predictions of ET, SH, BW, and GPP, while eight features are crucial for predicting AGB and BRe2t. Even though the XGBoost algorithm has an excellent ability to capture complex nonlinear relationships, it does not predict the PFT-competition-related variables of AGB and BRe2t well because the physical model cannot robustly predict coexisting PFTs due to the higher dimensionality of predicting PFT composition as compared to other ecosystem variables. Another important point worth mentioning is the small sample size of coexistence cases in Exp-CTR, with only 309 cases having BRe2t in the range of [0.1, 0.9], while the majority of cases are dominated by either early or late successional PFT. This limited sample size may not provide enough data to train the XGBoost surrogate model sufficiently for predicting BRe2t within the range of [0.1, 0.9]. Therefore, further studies are still needed to improve the emulation of PFT-competition-related variables. Other approaches that have been applied in VDMs but not specifically for PFT coexistence modeling, for example, the generalized likelihood uncertainty estimation (GLUE) approach (Zhang et al., 2022) and the Bayesian model emulation approach (Fer et al., 2018), could provide alternative ways. Furthermore, we suggest exploring other machine learning algorithms, such as Gaussian processes and neural network algorithms, which may be better suited for capturing nonlinear correlations and learning from sparse data.
Overall, our study presents a reproducible approach that utilizes machine learning to identify parameter values that improve model fidelity against observations and promote coexistence between plant functional types in vegetation demography models across diverse ecosystems. This approach has the potential to enhance the modeling of PFT coexistence in other ecosystems, such as the mixed conifer forests in Sierra Nevada, California (Buotte et al., 2021); Amazon forests subject to selective logging (Huang et al., 2020); and tropical forests with heterogeneous soils and subject to droughts in Panama (Cheng et al., 2021).
4.3 Trait tradeoffs between coexisting PFTs
Trait-related parameters show tradeoffs between early and late successional PFTs for the ELM-FATES-simulated coexisting experiments. The relative differences between the two PFTs in SLA, Vcmax, and WD complementarily coordinate with the relative difference in Mbk and Lleaf and hence avoid competitive exclusion (Fig. 9). These ELM-FATES reflected tradeoffs are consistent with the niche-based species coexistence mechanisms of environmental filtering and niche partitioning (Michalko and Pekár, 2015; Adler et al., 2013). On the one hand, in the coexisting cases, the relative differences between the two PFTs' parameters should not be considerable. For example, a large difference in SLA more likely favors the early cases (dashed green box in Fig. 9). This is related to environmental filtering in which coexisting species require some degree of convergence in strategy to survive and persist under given environmental conditions (Cadotte and Tucker, 2017; Thakur and Wright, 2017). On the other hand, some degree of differences should exist between the two PFTs' parameters in the coexisting cases. This is related to niche partitioning to ensure either differences in resource requirements or differences in tolerance to surrounding conditions (Kraft et al., 2015; Fowler et al., 2013). Phenomenological evidence has shown that functional trait variation promotes coexistence or increases species richness (Uriarte et al., 2010; Angert et al., 2009; Adler et al., 2006; Mason et al., 2012; Ben-Hur et al., 2012).
In our ELM-FATES simulations, the primary axis of competition for resources is light. The tradeoffs between the two PFTs' parameters differentiate their vertical competition in light absorption, which has been shown to strongly control tropical forest community composition (Farrior et al., 2016; Poorter et al., 2003). Even though the early PFT has a shallower rooting depth than the late PFT, there is no critical dry condition during our simulation period (i.e., corresponding to values of the water stress factor (BTRAN) close to 1.0 in Fig. S7). Therefore, competition for water resource access negligibly contributes to PFT coexistence in this study. Previous tropical studies also revealed these coexistence mechanisms. At a tropical forest site in eastern Ecuador, Kraft et al. (2008) found that concurring trees are often less ecologically similar, and both environmental filtering (different topographic habitats of ridgetops vs. valley) and niche differentiation simultaneously contribute to species coexistence. Swenson and Enquist (2009) also found that at small spatial scales in a tropical forest, most traits of coexisting species were under-dispersed, consistent with environmental filtering, while the seed mass and maximum height were over-dispersed, reflecting niche partitioning.
4.4 Limitations and further model development
Some limitations exist in our experiments. Niche partitioning is a critical aspect of promoting species coexistence, which is closely related to spatial heterogeneity, temporal heterogeneity, disturbances (e.g., nature enemy, fire), and resource partitioning (Adler et al., 2013). In our current ELM-FATES simulations, some processes that have been or are being developed in the model are not considered. These processes include nutrient limitation (Holm et al., 2020), fire disturbance (Fisher et al., 2015), subsurface lateral flow (Fang et al., 2022), and plant hydraulics (Chitra-Tarak et al., 2021; Li et al., 2021). Ignoring these processes could limit the potential of niche partitioning among PFTs in our ELM-FATES simulations. Topography has been recognized as an essential spatial heterogeneity factor for tropical forests, but it is not considered in ELM-FATES (Kraft et al., 2008; Costa et al., 2022). For example, Fang et al. (2022) coupled a three-dimensional hydrology model (ParFlow) with ELM-FATES and found that lateral flow plays a prominent role in governing aboveground biomass, and Cheng et al. (2021) also found a critical role for subsurface hydrology on coexistence. As these processes are added to the model, the reproducibility aspects of the XGBoost method to identify PFT combinations that match a broad range of criteria will be particularly important.
Lacking other features or processes could also affect PFT coexistence in the current FATES. For example, plant trait plasticity, the idea that plants can adjust their morphological and/or physiological traits to better adapt to the environment (Nicotra et al., 2010; Bloomfield et al., 2018; McDowell et al., 2022), is also not appropriately considered in FATES. Leaf traits such as Vcmax and SLA do vary vertically through the canopy in FATES via a prescribed relationship described by Lloyd et al. (2010). Liu and Ng (2019) found that the SLA of a desert shrub is significantly correlated with seasonal water availability. Additionally, FATES only considers the inter-PFT variance of functional traits (e.g., different Vcmax for early and late PFTs). However, studies revealed that trait variations commonly exist within and between species (Wright et al., 2005; Engemann et al., 2016; Meng et al., 2015; Dong et al., 2020; Siefert et al., 2015), and these play a vital role in maintaining plant diversity (Violle et al., 2012; Lu et al., 2017). Reproductive features that enhance competitive exclusion tendencies have been illustrated to affect coexistence (Maréchaux and Chave, 2017; Fisher et al., 2018). Hanbury-Brown et al. (2022) discussed the importance of the representation of forest regeneration, including improving parameters and algorithms for reproductive allocation, dispersal, seed survival and germination, environmental filtering in the seedling layer, and tree regeneration strategies adapted to wind, fire, and anthropogenic disturbance regimes. Besides, both growth–survival and stature–recruitment tradeoffs are critical to accurately predict successional patterns in tropical forest structure and competition (see details in Rüger et al., 2020), which should also be better considered in future model development. Furthermore, measured plant traits are increasingly available. For example, the TRY datasets (Kattge et al., 2020) can be used to improve the model process and parameterizations. Future studies into properly and adequately using these datasets to guide VDM parameterizations are advocated.
4.5 Enhancing VDM prediction with machine learning
We provide a brief overview of how machine learning can be applied to improve the modeling of plant dynamics, specifically in the context of vegetation demographic models. Firstly, ML can be used to derive trait parameter values. For instance, in this study, ML could be applied to replace the simple equations to derive the relationships between measured traits (Sect. 4.1). By integrating multiple datasets, including in situ measurements, atmospheric forcing, and remote sensing, ML could derive the spatial patterns and temporal variations in trait parameters for use in large-scale VDM modeling. Secondly, ML can be utilized to optimize parameters by developing surrogate models that emulate the relationships between the parameters and the VDM simulations and using the surrogate models to identify optimal parameter values. This application has demonstrated success in this study and previous studies (e.g., Tsai et al., 2021; Dagon et al., 2020; Watson-Parris et al., 2021). Another benefit of using ML in VDMs is the ability to develop benchmark datasets. For example, studies have successfully employed ML to derive AGB datasets for various ecosystems (Morais et al., 2021; Zhang et al., 2020; Li et al., 2020; da Bispo et al., 2020; Pham et al., 2020). These datasets can serve as benchmarks to evaluate the accuracy of VDM simulations. Lastly, ML can be used to replace semiempirical sub-models with only small theoretical bases in DGVMs (Reichstein et al., 2019). For example, accurately modeling wildfire using process-based wildfire models integrated in DGVMs remains challenging. However, ML-based wildfire models have shown advantages in accuracy and computational efficiency (Rodrigues and de la Riva, 2014; Jain et al., 2020; Sayad et al., 2019) and have the potential to be employed in Earth system models to improve wildfire simulations (e.g., Zhu et al., 2022).
In this study, we explored two possible solutions to improve PFT coexistence modeling in a cohort-based model (ELM-FATES): (1) using plant trait relationships established from field measurements and (2) using machine learning surrogate models to optimize trait parameter values. Three ensembles of ELM-FATES experiments were conducted over a tropical forest site at Manaus, Brazil. We found that considering the observed trait relationships (Exp-OBS) slightly improves the simulations of water (ET), energy (SH and BW), and carbon (GPP, AGB) when compared against observations but degrades the simulation of PFT coexistence. Based on Exp-CTR, the ML surrogate models were built to optimize the ELM-FATES parameters by integrating the observations (i.e., ET, SH, BW, GPP, and AGB) and PFT coexistence criteria. Exp-ML, with parameters selected by the ML surrogate models, vastly improves the simulation of PFT coexistence and also better reproduces the annual means and seasonal variations in ET, SH, BW, GPP, and the filed inventory of AGB. This study demonstrates the benefits of using machine learning models to improve the modeling of PFT coexistence in ELM-FATES, with important implications for modeling the response and feedback of ecosystem dynamics to climate change. Our results also suggest that the incorporation of additional mechanisms into ELM-FATES is essential for robust modeling of coexisting PFTs.
The ELM-FATES source code, surface and domain data, forcing data, and ML codes used in this study are all archived on Zenodo (https://doi.org/10.5281/zenodo.7730685; Li, 2022). The observational reference datasets of GPP, ET, SH, BW, and AGB are obtained from Holm et al. (2020). The forcing data are available from Oak Ridge National Laboratory Distributed Active Archive Center (ORNL DAAC), https://doi.org/10.3334/ORNLDAAC/1842 (Restrepo-Coupe et al., 2021).
The supplement related to this article is available online at: https://doi.org/10.5194/gmd-16-4017-2023-supplement.
LL and YF designed and conducted the experiments and analysis and drafted the manuscript. ZZ and MS contributed to the machine learning, experiment design, and improvement of the manuscript. LRL contributed to the interpretation and discussion of results and improvement of the manuscript. ML, CDK, JAH, RAF, NGM, and JC contributed to the dataset, data interpretation, discussion, and modification of the manuscript.
The contact author has declared that none of the authors has any competing interests.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This research was conducted at Pacific Northwest National Laboratory, operated for the U.S. Department of Energy by Battelle Memorial Institute under contract DE-AC05-76RL01830. This study was supported by the Department of Energy's (DOE) Office of Biological and Environmental Research as part of the Terrestrial Ecosystem Science program through the Next-Generation Ecosystem Experiments (NGEE)-Tropics project. Rosie A. Fisher acknowledges funding by the European Union's Horizon 2020 (H2020) research and innovation program under grant agreement nos. 101003536 (ESM2025 – Earth System Models for the Future) and 821003 (4C, Climate–Carbon Interactions in the Coming Century).
This research was supported by the U.S. Department of Energy, Office of Science (grant no. 71073).
This paper was edited by Klaus Klingmüller and reviewed by two anonymous referees.
Adler, P. B., HilleRisLambers, J., Kyriakidis, P. C., Guan, Q., and Levine, J. M.: Climate variability has a stabilizing effect on the coexistence of prairie grasses, P. Natl. Acad. Sci. USA, 103, 12793–12798, https://doi.org/10.1073/pnas.0600599103, 2006.
Adler, P. B., Fajardo, A., Kleinhesselink, A. R., and Kraft, N. J. B.: Trait-based tests of coexistence mechanisms, Ecol. Lett., 16, 1294–1306, https://doi.org/10.1111/ele.12157, 2013.
Angert, A. L., Huxman, T. E., Chesson, P., and Venable, D. L.: Functional tradeoffs determine species coexistence via the storage effect, P. Natl. Acad. Sci. USA, 106, 11641–11645, https://doi.org/10.1073/pnas.0904512106, 2009.
Antoniadis, A., Lambert-Lacroix, S., and Poggi, J.-M.: Random forests for global sensitivity analysis: A selective review, Reliab. Eng. Syst. Safe., 206, 107312, https://doi.org/10.1016/j.ress.2020.107312, 2020.
Bauman, D., Fortunel, C., Delhaye, G., Malhi, Y., Cernusak, L. A., Bentley, L. P., Rifai, S. W., Aguirre-Gutiérrez, J., Menor, I. O., Phillips, O. L., McNellis, B. E., Bradford, M., Laurance, S. G. W., Hutchinson, M. F., Dempsey, R., Santos-Andrade, P. E., Ninantay-Rivera, H. R., Paucar, J. R. C., and McMahon, S. M.: Tropical tree mortality has increased with rising atmospheric water stress, Nature, 608, 1–6, https://doi.org/10.1038/s41586-022-04737-7, 2022.
Ben-Hur, E., Fragman-Sapir, O., Hadas, R., Singer, A., and Kadmon, R.: Functional trade-offs increase species diversity in experimental plant communities, Ecol. Lett., 15, 1276–1282, https://doi.org/10.1111/j.1461-0248.2012.01850.x, 2012.
Berzaghi, F., Wright, I. J., Kramer, K., Oddou-Muratorio, S., Bohn, F. J., Reyer, C. P. O., Sabaté, S., Sanders, T. G. M., and Hartig, F.: Towards a New Generation of Trait-Flexible Vegetation Models, Trends Ecol. Evol., 35, 191–205, https://doi.org/10.1016/j.tree.2019.11.006, 2019.
Bloomfield, K. J., Cernusak, L. A., Eamus, D., Ellsworth, D. S., Prentice, I. C., Wright, I. J., Boer, M. M., Bradford, M. G., Cale, P., Cleverly, J., Egerton, J. J. G., Evans, B. J., Hayes, L. S., Hutchinson, M. F., Liddell, M. J., Macfarlane, C., Meyer, W. S., Prober, S. M., Togashi, H. F., Wardlaw, T., Zhu, L., and Atkin, O. K.: A continental-scale assessment of variability in leaf traits: Within species, across sites and between seasons, Funct. Ecol., 32, 1492–1506, https://doi.org/10.1111/1365-2435.13097, 2018.
Bonan, G. B.: Forests and Climate Change: Forcings, Feedbacks, and the Climate Benefits of Forests, Science, 320, 1444–1449, https://doi.org/10.1126/science.1155121, 2008.
Breiman, L.: Random Forests, Mach. Learn., 45, 5–32, https://doi.org/10.1023/a:1010933404324, 2001.
Brister, E., Newhouse, A. E., and Texas, C. E. P.: The University of North: Not the Same Old Chestnut: Rewilding Forests with Biotechnology, Environ. Ethics, 42, 149–167, https://doi.org/10.5840/enviroethics2020111614, 2020.
Buotte, P. C., Koven, C. D., Xu, C., Shuman, J. K., Goulden, M. L., Levis, S., Katz, J., Ding, J., Ma, W., Robbins, Z., and Kueppers, L. M.: Capturing functional strategies and compositional dynamics in vegetation demographic models, Biogeosciences, 18, 4473–4490, https://doi.org/10.5194/bg-18-4473-2021, 2021.
Cadotte, M. W. and Tucker, C. M.: Should Environmental Filtering be Abandoned?, Trends Ecol. Evol., 32, 429–437, https://doi.org/10.1016/j.tree.2017.03.004, 2017.
Cao, M. and Woodward, F. I.: Dynamic responses of terrestrial ecosystem carbon cycling to global climate change, Nature, 393, 249–252, https://doi.org/10.1038/30460, 1998.
Chen, T. and Guestrin, C.: XGBoost: A Scalable Tree Boosting System, Proc. 22nd Acm. Sigkdd. Int. Conf. Knowl. Discov. Data Min., 785–794, https://doi.org/10.1145/2939672.2939785, 2016.
Cheng, Y., Leung, L. R., Huang, M., Koven, C., Detto, M., Knox, R., Bisht, G., Bretfeld, M., and Fisher, R. A.: Modeling the joint effects of vegetation characteristics and soil properties on ecosystem dynamics in a Panama tropical forest, J. Adv. Model Earth. Sy., 14, e2021MS002603, https://doi.org/10.1029/2021ms002603, 2021.
Chitra-Tarak, R., Xu, C., Aguilar, S., Anderson-Teixeira, K. J., Chambers, J., Detto, M., Faybishenko, B., Fisher, R. A., Knox, R. G., Koven, C. D., Kueppers, L. M., Kunert, N., Kupers, S. J., McDowell, N. G., Newman, B. D., Paton, S. R., Pérez, R., Ruiz, L., Sack, L., Warren, J. M., Wolfe, B. T., Wright, C., Wright, S. J., Zailaa, J., and McMahon, S. M.: Hydraulically-vulnerable trees survive on deep-water access during droughts in a tropical forest, New Phytol., 231, 1798–1813, https://doi.org/10.1111/nph.17464, 2021.
Christoffersen, B. O., Gloor, M., Fauset, S., Fyllas, N. M., Galbraith, D. R., Baker, T. R., Kruijt, B., Rowland, L., Fisher, R. A., Binks, O. J., Sevanto, S., Xu, C., Jansen, S., Choat, B., Mencuccini, M., McDowell, N. G., and Meir, P.: Linking hydraulic traits to tropical forest function in a size-structured and trait-driven model (TFS v.1-Hydro), Geosci. Model Dev., 9, 4227–4255, https://doi.org/10.5194/gmd-9-4227-2016, 2016.
Costa, F. R. C., Schietti, J., Stark, S. C., and Smith, M. N.: The other side of tropical forest drought: do shallow water table regions of Amazonia act as large-scale hydrological refugia from drought?, New Phytol., 237, 714–733, https://doi.org/10.1111/nph.17914, 2022.
da Bispo, P. C., Rodríguez-Veiga, P., Zimbres, B., Miranda, S. do C. de, Cezare, C. H. G., Fleming, S., Baldacchino, F., Louis, V., Rains, D., Garcia, M., Espírito-Santo, F. D. B., Roitman, I., Pacheco-Pascagaza, A. M., Gou, Y., Roberts, J., Barrett, K., Ferreira, L. G., Shimbo, J. Z., Alencar, A., Bustamante, M., Woodhouse, I. H., Sano, E. E., Ometto, J. P., Tansey, K., and Balzter, H.: Woody Aboveground Biomass Mapping of the Brazilian Savanna with a Multi-Sensor and Machine Learning Approach, Remote Sens.-Basel, 12, 2685, https://doi.org/10.3390/rs12172685, 2020.
Dagon, K., Sanderson, B. M., Fisher, R. A., and Lawrence, D. M.: A machine learning approach to emulation and biophysical parameter estimation with the Community Land Model, version 5, Adv. Stat. Clim. Meteorol. Oceanogr., 6, 223–244, https://doi.org/10.5194/ascmo-6-223-2020, 2020.
Da Rocha, H. R., Manzi, A. O., Cabral, O. M., Miller, S. D., Goulden, M. L., Saleska, S. R., et al.: Patterns of water and heat flux across a biome gradient from tropical forest to savanna in Brazil, J. Geophys. Res.-Biogeo., 114, G00B12, https://doi.org/10.1029/2007JG000640, 2009.
Díaz, S., Kattge, J., Cornelissen, J. H. C., Wright, I. J., Lavorel, S., Dray, S., Reu, B., Kleyer, M., Wirth, C., Prentice, I. C., Garnier, E., Bönisch, G., Westoby, M., Poorter, H., Reich, P. B., Moles, A. T., Dickie, J., Gillison, A. N., Zanne, A. E., Chave, J., Wright, S. J., Sheremet'ev, S. N., Jactel, H., Baraloto, C., Cerabolini, B., Pierce, S., Shipley, B., Kirkup, D., Casanoves, F., Joswig, J. S., Günther, A., Falczuk, V., Rüger, N., Mahecha, M. D., and Gorné, L. D.: The global spectrum of plant form and function, Nature, 529, 167–171, https://doi.org/10.1038/nature16489, 2016.
Domingues, T. F., Berry, J. A., Martinelli, L. A., Ometto, J. P. H. B., and Ehleringer, J. R.: Parameterization of Canopy Structure and Leaf-Level Gas Exchange for an Eastern Amazonian Tropical Rain Forest (Tapajós National Forest, Pará, Brazil), Earth Interact., 9, 1–23, https://doi.org/10.1175/ei149.1, 2005.
Dong, N., Prentice, I. C., Wright, I. J., Evans, B. J., Togashi, H. F., Caddy-Retalic, S., McInerney, F. A., Sparrow, B., Leitch, E., and Lowe, A. J.: Components of leaf-trait variation along environmental gradients, New Phytol., 228, 82–94, https://doi.org/10.1111/nph.16558, 2020.
Duan, Q., Sorooshian, S., and Gupta, V.: Effective and efficient global optimization for conceptual rainfall-runoff models, Water Resour. Res., 28, 1015–1031, https://doi.org/10.1029/91wr02985, 1992.
Engemann, K., Sandel, B., Boyle, B., Enquist, B. J., Jørgensen, P. M., Kattge, J., McGill, B. J., Morueta-Holme, N., Peet, R. K., Spencer, N. J., Violle, C., Wiser, S. K., and Svenning, J.-C.: A plant growth form dataset for the New World, Ecology, 97, 3243–3243, https://doi.org/10.1002/ecy.1569, 2016.
Fang, Y., Leung, L. R., Duan, Z., Wigmosta, M. S., Maxwell, R. M., Chambers, J. Q., and Tomasella, J.: Influence of landscape heterogeneity on water available to tropical forests in an Amazonian catchment and implications for modeling drought response, J. Geophys. Res.-Atmos., 122, 8410–8426, https://doi.org/10.1002/2017jd027066, 2017.
Fang, Y., Leung, L. R., Koven, C. D., Bisht, G., Detto, M., Cheng, Y., McDowell, N., Muller-Landau, H., Wright, S. J., and Chambers, J. Q.: Modeling the topographic influence on aboveground biomass using a coupled model of hillslope hydrology and ecosystem dynamics, Geosci. Model Dev., 15, 7879–7901, https://doi.org/10.5194/gmd-15-7879-2022, 2022.
Farrior, C. E., Bohlman, S. A., Hubbell, S., and Pacala, S. W.: Dominance of the suppressed: Power-law size structure in tropical forests, Science, 351, 155–157, https://doi.org/10.1126/science.aad0592, 2016.
FATES Development Team: Technical Note for the Functionally Assembled Terrestrial Ecosystem Simulator (FATES) (v0.0.0), Zenodo [data set], https://doi.org/10.5281/zenodo.3517272, 2019.
Feeley, K. J., Davies, S. J., Ashton, P. S., Bunyavejchewin, S., Supardi, M. N. N., Kassim, A. R., Tan, S., and Chave, J.: The role of gap phase processes in the biomass dynamics of tropical forests, P. Roy. Soc. B, 274, 2857–2864, https://doi.org/10.1098/rspb.2007.0954, 2007.
Feigl, M., Lebiedzinski, K., Herrnegger, M., and Schulz, K.: Machine-learning methods for stream water temperature prediction, Hydrol. Earth Syst. Sci., 25, 2951–2977, https://doi.org/10.5194/hess-25-2951-2021, 2021.
Fer, I., Kelly, R., Moorcroft, P. R., Richardson, A. D., Cowdery, E. M., and Dietze, M. C.: Linking big models to big data: efficient ecosystem model calibration through Bayesian model emulation, Biogeosciences, 15, 5801–5830, https://doi.org/10.5194/bg-15-5801-2018, 2018.
Fisher, R., McDowell, N., Purves, D., Moorcroft, P., Sitch, S., Cox, P., Huntingford, C., Meir, P., and Woodward, F. I.: Assessing uncertainties in a second-generation dynamic vegetation model caused by ecological scale limitations, New Phytol., 187, 666–681, https://doi.org/10.1111/j.1469-8137.2010.03340.x, 2010.
Fisher, R. A., Muszala, S., Verteinstein, M., Lawrence, P., Xu, C., McDowell, N. G., Knox, R. G., Koven, C., Holm, J., Rogers, B. M., Spessa, A., Lawrence, D., and Bonan, G.: Taking off the training wheels: the properties of a dynamic vegetation model without climate envelopes, CLM4.5(ED), Geosci. Model Dev., 8, 3593–3619, https://doi.org/10.5194/gmd-8-3593-2015, 2015.
Fisher, R. A., Koven, C. D., Anderegg, W. R. L., Christoffersen, B. O., Dietze, M. C., Farrior, C. E., Holm, J. A., Hurtt, G. C., Knox, R. G., Lawrence, P. J., Lichstein, J. W., Longo, M., Matheny, A. M., Medvigy, D., Muller-Landau, H. C., Powell, T. L., Serbin, S. P., Sato, H., Shuman, J. K., Smith, B., Trugman, A. T., Viskari, T., Verbeeck, H., Weng, E., Xu, C., Xu, X., Zhang, T., and Moorcroft, P. R.: Vegetation demographics in Earth System Models: A review of progress and priorities, Global Change Biol., 24, 35–54, https://doi.org/10.1111/gcb.13910, 2018.
Foken, T.: The energy balance closure problem: An overview, Ecol. Appl., 18, 1351–1367, https://doi.org/10.1890/06-0922.1, 2008.
Foley, J. A., Prentice, I. C., Ramankutty, N., Levis, S., Pollard, D., Sitch, S., and Haxeltine, A.: An integrated biosphere model of land surface processes, terrestrial carbon balance, and vegetation dynamics, Global Biogeochem. Cy., 10, 603–628, https://doi.org/10.1029/96gb02692, 1996.
Fowler, D., Lessard, J.-P., and Sanders, N. J.: Niche filtering rather than partitioning shapes the structure of temperate forest ant communities, J. Animal Ecol., 83, 943–952, https://doi.org/10.1111/1365-2656.12188, 2013.
Friedman, J. H.: Greedy function approximation: A gradient boosting machine, Ann. Stat., 29, 1189–1232, https://doi.org/10.1214/aos/1013203451, 2001.
Fyllas, N. M., Gloor, E., Mercado, L. M., Sitch, S., Quesada, C. A., Domingues, T. F., Galbraith, D. R., Torre-Lezama, A., Vilanova, E., Ramírez-Angulo, H., Higuchi, N., Neill, D. A., Silveira, M., Ferreira, L., Aymard C., G. A., Malhi, Y., Phillips, O. L., and Lloyd, J.: Analysing Amazonian forest productivity using a new individual and trait-based model (TFS v.1), Geosci. Model Dev., 7, 1251–1269, https://doi.org/10.5194/gmd-7-1251-2014, 2014.
Gatti, L. V., Basso, L. S., Miller, J. B., Gloor, M., Domingues, L. G., Cassol, H. L. G., Tejada, G., Aragão, L. E. O. C., Nobre, C., Peters, W., Marani, L., Arai, E., Sanches, A. H., Corrêa, S. M., Anderson, L., Randow, C. V., Correia, C. S. C., Crispim, S. P., and Neves, R. A. L.: Amazonia as a carbon source linked to deforestation and climate change, Nature, 595, 388–393, https://doi.org/10.1038/s41586-021-03629-6, 2021.
Golaz, J., Caldwell, P. M., Roekel, L. P. V., Petersen, M. R., Tang, Q., Wolfe, J. D., Abeshu, G., Anantharaj, V., Asay-Davis, X. S., Bader, D. C., Baldwin, S. A., Bisht, G., Bogenschutz, P. A., Branstetter, M., Brunke, M. A., Brus, S. R., Burrows, S. M., Cameron-Smith, P. J., Donahue, A. S., Deakin, M., Easter, R. C., Evans, K. J., Feng, Y., Flanner, M., Foucar, J. G., Fyke, J. G., Griffin, B. M., Hannay, C., Harrop, B. E., Hoffman, M. J., Hunke, E. C., Jacob, R. L., Jacobsen, D. W., Jeffery, N., Jones, P. W., Keen, N. D., Klein, S. A., Larson, V. E., Leung, L. R., Li, H., Lin, W., Lipscomb, W. H., Ma, P., Mahajan, S., Maltrud, M. E., Mametjanov, A., McClean, J. L., McCoy, R. B., Neale, R. B., Price, S. F., Qian, Y., Rasch, P. J., Eyre, J. E. J. R., Riley, W. J., Ringler, T. D., Roberts, A. F., Roesler, E. L., Salinger, A. G., Shaheen, Z., Shi, X., Singh, B., Tang, J., Taylor, M. A., Thornton, P. E., Turner, A. K., Veneziani, M., Wan, H., Wang, H., Wang, S., Williams, D. N., Wolfram, P. J., Worley, P. H., Xie, S., Yang, Y., Yoon, J., Zelinka, M. D., Zender, C. S., Zeng, X., Zhang, C., Zhang, K., Zhang, Y., Zheng, X., Zhou, T., and Zhu, Q.: The DOE E3SM Coupled Model Version 1: Overview and Evaluation at Standard Resolution, J. Adv. Model Earth Sy., 11, 2089–2129, https://doi.org/10.1029/2018ms001603, 2019.
Hanbury-Brown, A. R., Ward, R. E., and Kueppers, L. M.: Forest regeneration within Earth system models: current process representations and ways forward, New Phytol., 235, 20–40, https://doi.org/10.1111/nph.18131, 2022.
Haverd, V., Smith, B., Cook, G. D., Briggs, P. R., Nieradzik, L., Roxburgh, S. H., Liedloff, A., Meyer, C. P., and Canadell, J. G.: A stand-alone tree demography and landscape structure module for Earth system models, Geophys. Res. Lett., 40, 5234–5239, https://doi.org/10.1002/grl.50972, 2013.
He, X., Liu, S., Xu, T., Yu, K., Gentine, P., Zhang, Z., Xu, Z., Jiao, D., and Wu, D.: Improving predictions of evapotranspiration by integrating multi-source observations and land surface model, Agr. Water Manage., 272, 107827, https://doi.org/10.1016/j.agwat.2022.107827, 2022.
Holm, J. A., Knox, R. G., Zhu, Q., Fisher, R. A., Koven, C. D., Lima, A. J. N., Riley, W. J., Longo, M., Negrón-Juárez, R. I., Araujo, A. C., Kueppers, L. M., Moorcroft, P. R., Higuchi, N., and Chambers, J. Q.: The Central Amazon Biomass Sink Under Current and Future Atmospheric CO2: Predictions From Big-Leaf and Demographic Vegetation Models, J. Geophys. Res.-Biogeo., 125, e2019JG005500, https://doi.org/10.1029/2019jg005500, 2020.
Huang, M., Xu, Y., Longo, M., Keller, M., Knox, R. G., Koven, C. D., and Fisher, R. A.: Assessing impacts of selective logging on water, energy, and carbon budgets and ecosystem dynamics in Amazon forests using the Functionally Assembled Terrestrial Ecosystem Simulator, Biogeosciences, 17, 4999–5023, https://doi.org/10.5194/bg-17-4999-2020, 2020.
Hubau, W., Lewis, S. L., Phillips, O. L., Affum-Baffoe, K., Beeckman, H., Cuní-Sanchez, A., Daniels, A. K., Ewango, C. E. N., Fauset, S., Mukinzi, J. M., Sheil, D., Sonké, B., Sullivan, M. J. P., Sunderland, T. C. H., Taedoumg, H., Thomas, S. C., White, L. J. T., Abernethy, K. A., Adu-Bredu, S. A., Amani, C. A., Baker, T. R., Banin, L. F., Baya, F., Begne, S. K., Bennett, A. C., Benedet, F., Bitariho, R., Bocko, Y. E., Boeckx, P., Boundja, P., Brienen, R. J. W., Brncic, T., Chezeaux, E., Chuyong, G. B., Clark, C. J., Collins, M., Comiskey, J. A., Coomes, D. A., Dargie, G. C., de Haulleville, T., Djuikouo Kamdem, M. N., Doucet, J. L., Esquivel-Muelbert, A., Feldpausch, T. R., Fofanah, A., Foli, E. G., Gilpin, M., Gloor, E., Gonmadje, C., Gourlet-Fleury, S., Hall, J. S., Hamilton, A. C., Harris, D. J., Hart, T. B., Hockemba, M. B. N., Hladik, A., Ifo, S. A., Jeffery, K. J., Jucker, T., Kasongo Yakusu, E., Kearsley, E., Kenfack, D., Koch, A., Leal, M. E., Levesley, A., Lindsell, J. A., Lisingo, J., Lopez-Gonzalez, G., Lovett, J. C., Makana, J.-R., Malhi, Y., Marshall, A. R., Martin, J., Martin, E. H., Mbayu, F. M., Medjibe, V. P., Mihindou, V., Mitchard, E. T. A., Moore, S., Munishi, P. K. T., Nssi Bengone, N., Ojo, L., Evouna Ondo, F., Peh, K. S.-H., Pickavance, G. C., Poulsen, A. D., Poulsen, J. R., Qie, L., Reitsma, J., Rovero, F., Swaine, M. D., Talbot, J., Taplin, J., Taylor, D. M., Thomas, D. W., Toirambe, B., Tshibamba Mukendi, J., Tuagben, D., Umunay, P. M., van der Heijden, G. M. F., Verbeeck, H., Vleminckx, J., Willcock, S., Wöll, H., Woods, J. T., and Zemagho, L.: Asynchronous carbon sink saturation in African and Amazonian tropical forests, Nature, 579, 80–87, https://doi.org/10.1038/s41586-020-2035-0, 2020.
Hurtt, G. C., Moorcroft, PauL. R., And, S. W. P., and Levin, S. A.: Terrestrial models and global change: challenges for the future, Global Change Biol., 4, 581–590, https://doi.org/10.1046/j.1365-2486.1998.t01-1-00203.x, 1998.
Jain, P., Coogan, S. C. P., Subramanian, S. G., Crowley, M., Taylor, S., and Flannigan, M. D.: A review of machine learning applications in wildfire science and management, Arxiv, https://doi.org/10.48550/arxiv.2003.00646, 2020.
Jonard, M., André, F., de Coligny, F., de Wergifosse, L., Beudez, N., Davi, H., Ligot, G., Ponette, Q., and Vincke, C.: HETEROFOR 1.0: a spatially explicit model for exploring the response of structurally complex forests to uncertain future conditions – Part 1: Carbon fluxes and tree dimensional growth, Geosci. Model Dev., 13, 905–935, https://doi.org/10.5194/gmd-13-905-2020, 2020.
Jung, M., Koirala, S., Weber, U., Ichii, K., Gans, F., Camps-Valls, G., Papale, D., Schwalm, C., Tramontana, G., and Reichstein, M.: The FLUXCOM ensemble of global land-atmosphere energy fluxes., Sci. Data, 6, 74, https://doi.org/10.1038/s41597-019-0076-8, 2019.
Kattge, J., Bönisch, G., Díaz, S., et al.: TRY plant trait database – enhanced coverage and open access, Global Change Biol., 26, 119–188, https://doi.org/10.1111/gcb.14904, 2020.
Koven, C. D., Knox, R. G., Fisher, R. A., Chambers, J. Q., Christoffersen, B. O., Davies, S. J., Detto, M., Dietze, M. C., Faybishenko, B., Holm, J., Huang, M., Kovenock, M., Kueppers, L. M., Lemieux, G., Massoud, E., McDowell, N. G., Muller-Landau, H. C., Needham, J. F., Norby, R. J., Powell, T., Rogers, A., Serbin, S. P., Shuman, J. K., Swann, A. L. S., Varadharajan, C., Walker, A. P., Wright, S. J., and Xu, C.: Benchmarking and parameter sensitivity of physiological and vegetation dynamics using the Functionally Assembled Terrestrial Ecosystem Simulator (FATES) at Barro Colorado Island, Panama, Biogeosciences, 17, 3017–3044, https://doi.org/10.5194/bg-17-3017-2020, 2020.
Kraft, N. J. B., Valencia, R., and Ackerly, D. D.: Functional Traits and Niche-Based Tree Community Assembly in an Amazonian Forest, Science, 322, 580–582, https://doi.org/10.1126/science.1160662, 2008.
Kraft, N. J. B., Adler, P. B., Godoy, O., James, E. C., Fuller, S., and Levine, J. M.: Community assembly, coexistence and the environmental filtering metaphor, Funct. Ecol., 29, 592–599, https://doi.org/10.1111/1365-2435.12345, 2015.
Lambert, M. S. A., Tang, H., Aas, K. S., Stordal, F., Fisher, R. A., Fang, Y., Ding, J., and Parmentier, F.-J. W.: Inclusion of a cold hardening scheme to represent frost tolerance is essential to model realistic plant hydraulics in the Arctic–boreal zone in CLM5.0-FATES-Hydro, Geosci. Model Dev., 15, 8809–8829, https://doi.org/10.5194/gmd-15-8809-2022, 2022.
Lawrence, D. M., Fisher, R. A., Koven, C. D., Oleson, K. W., Swenson, S. C., Bonan, G., Collier, N., Ghimire, B., Kampenhout, L., Kennedy, D., Kluzek, E., Lawrence, P. J., Li, F., Li, H., Lombardozzi, D., Riley, W. J., Sacks, W. J., Shi, M., Vertenstein, M., Wieder, W. R., Xu, C., Ali, A. A., Badger, A. M., Bisht, G., Broeke, M., Brunke, M. A., Burns, S. P., Buzan, J., Clark, M., Craig, A., Dahlin, K., Drewniak, B., Fisher, J. B., Flanner, M., Fox, A. M., Gentine, P., Hoffman, F., Keppel-Aleks, G., Knox, R., Kumar, S., Lenaerts, J., Leung, L. R., Lipscomb, W. H., Lu, Y., Pandey, A., Pelletier, J. D., Perket, J., Randerson, J. T., Ricciuto, D. M., Sanderson, B. M., Slater, A., Subin, Z. M., Tang, J., Thomas, R. Q., Martin, M. V., and Zeng, X.: The Community Land Model Version 5: Description of New Features, Benchmarking, and Impact of Forcing Uncertainty, J. Adv. Model Earth Sy., 11, 4245–4287, https://doi.org/10.1029/2018ms001583, 2019.
Leung, L. R., Bader, D. C., Taylor, M. A., and McCoy, R. B.: An Introduction to the E3SM Special Collection: Goals, Science Drivers, Development, and Analysis, J. Adv. Model Earth Sy., 12, e2019MS001821, https://doi.org/10.1029/2019ms001821, 2020.
Li, L.: A machine learning approach targeting parameter estimation for plant functional type coexistence modeling using ELM-FATES, Zenodo [code], https://doi.org/10.5281/zenodo.7730685, 2022.
Li, L., Yang, Z., Matheny, A. M., Zheng, H., Swenson, S. C., Lawrence, D. M., Barlage, M., Yan, B., McDowell, N. G., and Leung, L. R.: Representation of Plant Hydraulics in the Noah-MP Land Surface Model: Model Development and Multiscale Evaluation, J. Adv. Model Earth Sy., 13, e2020MS002214, https://doi.org/10.1029/2020ms002214, 2021.
Li, Y., Li, M., Li, C., and Liu, Z.: Forest aboveground biomass estimation using Landsat 8 and Sentinel-1A data with machine learning algorithms, Sci. Rep.-UK, 10, 9952, https://doi.org/10.1038/s41598-020-67024-3, 2020.
Liu, S. and Ng, G.-H. C.: A data-conditioned stochastic parameterization of temporal plant trait variability in an ecohydrological model and the potential for plasticity, Agr. Forest Meteorol., 274, 184–194, https://doi.org/10.1016/j.agrformet.2019.05.005, 2019.
Lloyd, J., Patiño, S., Paiva, R. Q., Nardoto, G. B., Quesada, C. A., Santos, A. J. B., Baker, T. R., Brand, W. A., Hilke, I., Gielmann, H., Raessler, M., Luizão, F. J., Martinelli, L. A., and Mercado, L. M.: Optimisation of photosynthetic carbon gain and within-canopy gradients of associated foliar traits for Amazon forest trees, Biogeosciences, 7, 1833–1859, https://doi.org/10.5194/bg-7-1833-2010, 2010.
Longo, M., Knox, R. G., Medvigy, D. M., Levine, N. M., Dietze, M. C., Kim, Y., Swann, A. L. S., Zhang, K., Rollinson, C. R., Bras, R. L., Wofsy, S. C., and Moorcroft, P. R.: The biophysics, ecology, and biogeochemistry of functionally diverse, vertically and horizontally heterogeneous ecosystems: the Ecosystem Demography model, version 2.2 – Part 1: Model description, Geosci. Model Dev., 12, 4309–4346, https://doi.org/10.5194/gmd-12-4309-2019, 2019.
Longo, M., Saatchi, S., Keller, M., Bowman, K., Ferraz, A., Moorcroft, P. R., Morton, D. C., Bonal, D., Brando, P., Burban, B., Derroire, G., dos-Santos, M. N., Meyer, V., Saleska, S., Trumbore, S., and Vincent, G.: Impacts of Degradation on Water, Energy, and Carbon Cycling of the Amazon Tropical Forests, J. Geophys. Res.-Biogeo., 125, e2020JG005677, https://doi.org/10.1029/2020jg005677, 2020.
Lu, X., Wang, Y., Wright, I. J., Reich, P. B., Shi, Z., and Dai, Y.: Incorporation of plant traits in a land surface model helps explain the global biogeographical distribution of major forest functional types, Global Ecol. Biogeogr., 26, 304–317, https://doi.org/10.1111/geb.12535, 2017.
Lundberg, S. and Lee, S.-I.: A Unified Approach to Interpreting Model Predictions, Arxiv, https://doi.org/10.48550/arXiv.1705.07874, 2017.
Lundberg, S. M., Nair, B., Vavilala, M. S., Horibe, M., Eisses, M. J., Adams, T., Liston, D. E., Low, D. K.-W., Newman, S.-F., Kim, J., and Lee, S.-I.: Explainable machine-learning predictions for the prevention of hypoxaemia during surgery, Nat. Biomed. Eng., 2, 749–760, https://doi.org/10.1038/s41551-018-0304-0, 2018.
Lundberg, S. M., Erion, G., Chen, H., DeGrave, A., Prutkin, J. M., Nair, B., Katz, R., Himmelfarb, J., Bansal, N., and Lee, S.-I.: From local explanations to global understanding with explainable AI for trees, Nat. Mach. Intell., 2, 56–67, https://doi.org/10.1038/s42256-019-0138-9, 2020.
Ma, L., Hurtt, G., Ott, L., Sahajpal, R., Fisk, J., Lamb, R., Tang, H., Flanagan, S., Chini, L., Chatterjee, A., and Sullivan, J.: Global evaluation of the Ecosystem Demography model (ED v3.0), Geosci. Model Dev., 15, 1971–1994, https://doi.org/10.5194/gmd-15-1971-2022, 2022.
Maréchaux, I. and Chave, J.: An individual-based forest model to jointly simulate carbon and tree diversity in Amazonia: description and applications, Ecol. Monogr., 87, 632–664, https://doi.org/10.1002/ecm.1271, 2017.
Martín Belda, D., Anthoni, P., Wårlind, D., Olin, S., Schurgers, G., Tang, J., Smith, B., and Arneth, A.: LPJ-GUESS/LSMv1.0: a next-generation land surface model with high ecological realism, Geosci. Model Dev., 15, 6709–6745, https://doi.org/10.5194/gmd-15-6709-2022, 2022.
Mason, N. W. H., Richardson, S. J., Peltzer, D. A., Bello, F. de, Wardle, D. A., and Allen, R. B.: Changes in coexistence mechanisms along a long-term soil chronosequence revealed by functional trait diversity: Functional diversity along ecological gradients, J. Ecol., 100, 678–689, https://doi.org/10.1111/j.1365-2745.2012.01965.x, 2012.
McDowell, N. G., Allen, C. D., Anderson-Teixeira, K., Aukema, B. H., Bond-Lamberty, B., Chini, L., Clark, J. S., Dietze, M., Grossiord, C., Hanbury-Brown, A., Hurtt, G. C., Jackson, R. B., Johnson, D. J., Kueppers, L., Lichstein, J. W., Ogle, K., Poulter, B., Pugh, T. A. M., Seidl, R., Turner, M. G., Uriarte, M., Walker, A. P., and Xu, C.: Pervasive shifts in forest dynamics in a changing world, Science, 368, eaaz9463, https://doi.org/10.1126/science.aaz9463, 2020.
McDowell, N. G., Sapes, G., Pivovaroff, A., Adams, H. D., Allen, C. D., Anderegg, W. R. L., Arend, M., Breshears, D. D., Brodribb, T., Choat, B., Cochard, H., Cáceres, M. D., Kauwe, M. G. D., Grossiord, C., Hammond, W. M., Hartmann, H., Hoch, G., Kahmen, A., Klein, T., Mackay, D. S., Mantova, M., Martínez-Vilalta, J., Medlyn, B. E., Mencuccini, M., Nardini, A., Oliveira, R. S., Sala, A., Tissue, D. T., Torres-Ruiz, J. M., Trowbridge, A. M., Trugman, A. T., Wiley, E., and Xu, C.: Mechanisms of woody-plant mortality under rising drought, CO2 and vapour pressure deficit, Nat. Rev. Earth Environ., 3, 294–308, https://doi.org/10.1038/s43017-022-00272-1, 2022.
Mckay, M. D., Beckman, R. J., and Conover, W. J.: A Comparison of Three Methods for Selecting Values of Input Variables in the Analysis of Output From a Computer Code, Technometrics, 42, 55–61, https://doi.org/10.1080/00401706.2000.10485979, 2000.
McMahon, S. M., Harrison, S. P., Armbruster, W. S., Bartlein, P. J., Beale, C. M., Edwards, M. E., Kattge, J., Midgley, G., Morin, X., and Prentice, I. C.: Improving assessment and modelling of climate change impacts on global terrestrial biodiversity, Trends Ecol. Evol., 26, 249–259, https://doi.org/10.1016/j.tree.2011.02.012, 2011.
Medvigy, D., Wofsy, S. C., Munger, J. W., Hollinger, D. Y., and Moorcroft, P. R.: Mechanistic scaling of ecosystem function and dynamics in space and time: Ecosystem Demography model version 2, J. Geophys. Res.-Biogeo., 114, G01002, https://doi.org/10.1029/2008jg000812, 2009.
Meng, T.-T., Wang, H., Harrison, S. P., Prentice, I. C., Ni, J., and Wang, G.: Responses of leaf traits to climatic gradients: adaptive variation versus compositional shifts, Biogeosciences, 12, 5339–5352, https://doi.org/10.5194/bg-12-5339-2015, 2015.
Michalko, R. and Pekár, S.: Niche partitioning and niche filtering jointly mediate the coexistence of three closely related spider species (Araneae, Philodromidae), Ecol. Entomol., 40, 22–33, https://doi.org/10.1111/een.12149, 2015.
Mitchard, E. T. A.: The tropical forest carbon cycle and climate change, Nature, 559, 527–534, https://doi.org/10.1038/s41586-018-0300-2, 2018.
Moorcroft, P. R.: Recent advances in ecosystem-atmosphere interactions: an ecological perspective, P. Roy. Soc. Lond. B, 270, 1215–1227, https://doi.org/10.1098/rspb.2002.2251, 2003.
Moorcroft, P. R., Hurtt, G. C., and Pacala, S. W.: A Method for Scaling Vegetation Dynamics: The Ecosystem Demography Model (ED), Ecol. Monogr., 71, 557, https://doi.org/10.2307/3100036, 2001.
Mora, C., Tittensor, D. P., Adl, S., Simpson, A. G. B., and Worm, B.: How Many Species Are There on Earth and in the Ocean?, Plos Biol., 9, e1001127, https://doi.org/10.1371/journal.pbio.1001127, 2011.
Morais, T. G., Teixeira, R. F. M., Figueiredo, M., and Domingos, T.: The use of machine learning methods to estimate aboveground biomass of grasslands: A review, Ecol. Indic., 130, 108081, https://doi.org/10.1016/j.ecolind.2021.108081, 2021.
Nearing, G. S., Kratzert, F., Sampson, A. K., Pelissier, C. S., Klotz, D., Frame, J. M., Prieto, C., and Gupta, H. V.: What Role Does Hydrological Science Play in the Age of Machine Learning?, Water Resour. Res., 57, e2020WR028091, https://doi.org/10.1029/2020wr028091, 2021.
Nicotra, A. B., Atkin, O. K., Bonser, S. P., Davidson, A. M., Finnegan, E. J., Mathesius, U., Poot, P., Purugganan, M. D., Richards, C. L., Valladares, F., and van Kleunen, M.: Plant phenotypic plasticity in a changing climate, Trends Plant Sci., 15, 684–692, https://doi.org/10.1016/j.tplants.2010.09.008, 2010.
Oliveira, R. S., Eller, C. B., Barros, F. de V., Hirota, M., Brum, M., and Bittencourt, P.: Linking plant hydraulics and the fast–slow continuum to understand resilience to drought in tropical ecosystems, New Phytol., 230, 904–923, https://doi.org/10.1111/nph.17266, 2021.
Padarian, J., McBratney, A. B., and Minasny, B.: Game theory interpretation of digital soil mapping convolutional neural networks, SOIL, 6, 389–397, https://doi.org/10.5194/soil-6-389-2020, 2020.
Pal, A., Mahajan, S., and Norman, M. R.: Using Deep Neural Networks as Cost-Effective Surrogate Models for Super-Parameterized E3SM Radiative Transfer, Geophys. Res. Lett., 46, 6069–6079, https://doi.org/10.1029/2018gl081646, 2019.
Peatier, S., Sanderson, B. M., Terray, L., and Roehrig, R.: Investigating Parametric Dependence of Climate Feedbacks in the Atmospheric Component of CNRM-CM6-1, Geophys. Res. Lett., 49, e2021GL095084, https://doi.org/10.1029/2021gl095084, 2022.
Pham, T. D., Yokoya, N., Xia, J., Ha, N. T., Le, N. N., Nguyen, T. T. T., Dao, T. H., Vu, T. T. P., Pham, T. D., and Takeuchi, W.: Comparison of Machine Learning Methods for Estimating Mangrove Above-Ground Biomass Using Multiple Source Remote Sensing Data in the Red River Delta Biosphere Reserve, Vietnam, Remote Sens.-Basel, 12, 1334, https://doi.org/10.3390/rs12081334, 2020.
Piao, S., Wang, X., Wang, K., Li, X., Bastos, A., Canadell, J. G., Ciais, P., Friedlingstein, P., and Sitch, S.: Interannual variation of terrestrial carbon cycle: Issues and perspectives, Global Change Biol., 26, 300–318, https://doi.org/10.1111/gcb.14884, 2020.
Poorter, L., Bongers, F., Sterck, F. J., and Wöll, H.: Architecture of 53 rain forest tree species differing in adult stature and shade tolerance, 84, 602–608, https://doi.org/10.1890/0012-9658(2003)084[0602:aorfts]2.0.co;2, 2003.
Purves, D. W., Lichstein, J. W., Strigul, N., and Pacala, S. W.: Predicting and understanding forest dynamics using a simple tractable model, P. Natl. Acad. Sci. USA, 105, 17018–17022, https://doi.org/10.1073/pnas.0807754105, 2008.
Reich, P. B.: The world-wide “fast–slow” plant economics spectrum: a traits manifesto, J. Ecol., 102, 275–301, https://doi.org/10.1111/1365-2745.12211, 2014.
Reichstein, M., Camps-Valls, G., Stevens, B., Jung, M., Denzler, J., Carvalhais, N., and Prabhat: Deep learning and process understanding for data-driven Earth system science, Nature, 566, 195–204, https://doi.org/10.1038/s41586-019-0912-1, 2019.
Restrepo-Coupe, N., da Rocha, H. R., Hutyra, L. R., de Araujo, A. C., Borma, L. S., Christoffersen, B., Cabral, O., de Camargo, P. B., Cardoso, F. L., Costa, A. C. L., Fitzjarrald, D. R. Goulden,, M. L., Kruijt, B., Maia, J. M. F., Malhi, Y. S., Manzi, A. O., Miller, S. D., Nobre, A. D., von Randow, C., Abreu Safaj, L. D., Sakai, R. K., Tota, J., Wofsy, S. C., Zanchi, F. B., and Saleska, S. R.: LBA-ECO CD-32 Flux Tower Network Data Compilation, Brazilian Amazon: 1999–2006, V2, ORNL DAAC, Oak Ridge, Tennessee, USA [data set], https://doi.org/10.3334/ORNLDAAC/1842, 2021.
Rodrigues, M. and de la Riva, J.: An insight into machine-learning algorithms to model human-caused wildfire occurrence, Environ Modell Softw, 57, 192–201, https://doi.org/10.1016/j.envsoft.2014.03.003, 2014.
Rouholahnejad, E., Abbaspour, K. C., Vejdani, M., Srinivasan, R., Schulin, R., and Lehmann, A.: A parallelization framework for calibration of hydrological models, Environ. Modell. Softw., 31, 28–36, https://doi.org/10.1016/j.envsoft.2011.12.001, 2012.
Rüger, N., Condit, R., Dent, D. H., DeWalt, S. J., Hubbell, S. P., Lichstein, J. W., Lopez, O. R., Wirth, C., and Farrior, C. E.: Demographic trade-offs predict tropical forest dynamics, Science, 368, 165–168, https://doi.org/10.1126/science.aaz4797, 2020.
Sakschewski, B., Bloh, W., Boit, A., Rammig, A., Kattge, J., Poorter, L., Peñuelas, J., and Thonicke, K.: Leaf and stem economics spectra drive diversity of functional plant traits in a dynamic global vegetation model, Global Change Biol., 21, 2711–2725, https://doi.org/10.1111/gcb.12870, 2015.
Sakschewski, B., Bloh, W. von, Boit, A., Poorter, L., Peña-Claros, M., Heinke, J., Joshi, J., and Thonicke, K.: Resilience of Amazon forests emerges from plant trait diversity, Nat. Clim. Change, 6, 1032–1036, https://doi.org/10.1038/nclimate3109, 2016.
Sato, H., Itoh, A., and Kohyama, T.: SEIB–DGVM: A new Dynamic Global Vegetation Model using a spatially explicit individual-based approach, Ecol. Model., 200, 279–307, https://doi.org/10.1016/j.ecolmodel.2006.09.006, 2007.
Sawada, Y.: Machine Learning Accelerates Parameter Optimization and Uncertainty Assessment of a Land Surface Model, J. Geophys. Res.-Atmos., 125, e2020JD032688, https://doi.org/10.1029/2020jd032688, 2020.
Sayad, Y. O., Mousannif, H., and Moatassime, H. A.: Predictive modeling of wildfires: A new dataset and machine learning approach, Fire Safety J., 104, 130–146, https://doi.org/10.1016/j.firesaf.2019.01.006, 2019.
Scheiter, S., Langan, L., and Higgins, S. I.: Next‐generation dynamic global vegetation models: learning from community ecology, New Phytologist, 198, 957–969, https://doi.org/10.1111/nph.12210, 2013.
Shen, C.: A Transdisciplinary Review of Deep Learning Research and Its Relevance for Water Resources Scientists, Water Resour. Res., 54, 8558–8593, https://doi.org/10.1029/2018wr022643, 2018.
Siefert, A., Violle, C., Chalmandrier, L., Albert, C. H., Taudiere, A., Fajardo, A., Aarssen, L. W., Baraloto, C., Carlucci, M. B., Cianciaruso, M. V., Dantas, V. L., Bello, F., Duarte, L. D. S., Fonseca, C. R., Freschet, G. T., Gaucherand, S., Gross, N., Hikosaka, K., Jackson, B., Jung, V., Kamiyama, C., Katabuchi, M., Kembel, S. W., Kichenin, E., Kraft, N. J. B., Lagerström, A., Bagousse-Pinguet, Y. L., Li, Y., Mason, N., Messier, J., Nakashizuka, T., Overton, J. McC., Peltzer, D. A., Pérez-Ramos, I. M., Pillar, V. D., Prentice, H. C., Richardson, S., Sasaki, T., Schamp, B. S., Schöb, C., Shipley, B., Sundqvist, M., Sykes, M. T., Vandewalle, M., and Wardle, D. A.: A global meta-analysis of the relative extent of intraspecific trait variation in plant communities, Ecol. Lett., 18, 1406–1419, https://doi.org/10.1111/ele.12508, 2015.
Sit, M., Demiray, B. Z., Xiang, Z., Ewing, G. J., Sermet, Y., and Demir, I.: A comprehensive review of deep learning applications in hydrology and water resources, Water Sci. Technol., 82, 2635–2670, https://doi.org/10.2166/wst.2020.369, 2020.
Sitch, S., Smith, B., Prentice, I. C., Arneth, A., Bondeau, A., Cramer, W., Kaplan, J. O., Levis, S., Lucht, W., Sykes, M. T., Thonicke, K., and Venevsky, S.: Evaluation of ecosystem dynamics, plant geography and terrestrial carbon cycling in the LPJ dynamic global vegetation model: LPJ DYNAMIC GLOBAL VEGETATION MODEL, Global Change Biol., 9, 161–185, https://doi.org/10.1046/j.1365-2486.2003.00569.x, 2003.
Snell, R. S., Huth, A., Nabel, J. E. M. S., Bocedi, G., Travis, J. M. J., Gravel, D., Bugmann, H., Gutiérrez, A. G., Hickler, T., Higgins, S. I., Reineking, B., Scherstjanoi, M., Zurbriggen, N., and Lischke, H.: Using dynamic vegetation models to simulate plant range shifts, Ecography, 37, 1184–1197, https://doi.org/10.1111/ecog.00580, 2014.
Snoek, J., Larochelle, H., and Adams, R. P.: Practical Bayesian Optimization of Machine Learning Algorithms, Arxiv, https://doi.org/10.48550/arXiv.1206.2944, 2012.
Stark, S. C., Leitold, V., Wu, J. L., Hunter, M. O., de Castilho, C. V., Costa, F. R. C., McMahon, S. M., Parker, G. G., Shimabukuro, M. T., Lefsky, M. A., Keller, M., Alves, L. F., Schietti, J., Shimabukuro, Y. E., Brandão, D. O., Woodcock, T. K., Higuchi, N., de Camargo, P. B., de Oliveira, R. C., Saleska, S. R., and Chave, J.: Amazon forest carbon dynamics predicted by profiles of canopy leaf area and light environment, Ecol. Lett., 15, 1406–1414, https://doi.org/10.1111/j.1461-0248.2012.01864.x, 2012.
Strigul, N., Pristinski, D., Purves, D., Dushoff, J., and Pacala, S.: Scaling from trees to forests: tractable macroscopic equations for forest dynamics, Ecol. Monogr., 78, 523–545, https://doi.org/10.1890/08-0082.1, 2008.
Swenson, N. G. and Enquist, B. J.: Opposing assembly mechanisms in a Neotropical dry forest: implications for phylogenetic and functional community ecology, Ecology, 90, 2161–2170, https://doi.org/10.1890/08-1025.1, 2009.
Thakur, M. P. and Wright, A. J.: Environmental Filtering, Niche Construction, and Trait Variability: The Missing Discussion, Trends Ecol. Evol., 32, 884–886, https://doi.org/10.1016/j.tree.2017.09.014, 2017.
Tsai, W.-P., Feng, D., Pan, M., Beck, H., Lawson, K., Yang, Y., Liu, J., and Shen, C.: From calibration to parameter learning: Harnessing the scaling effects of big data in geoscientific modeling, Nat. Commun., 12, 5988, https://doi.org/10.1038/s41467-021-26107-z, 2021.
Uriarte, M., Swenson, N. G., Chazdon, R. L., Comita, L. S., Kress, W. J., Erickson, D., Forero-Montaña, J., Zimmerman, J. K., and Thompson, J.: Trait similarity, shared ancestry and the structure of neighbourhood interactions in a subtropical wet forest: implications for community assembly, Ecol. Lett., 13, 1503–1514, https://doi.org/10.1111/j.1461-0248.2010.01541.x, 2010.
Violle, C., Enquist, B. J., McGill, B. J., Jiang, L., Albert, C. H., Hulshof, C., Jung, V., and Messier, J.: The return of the variance: intraspecific variability in community ecology, Trends Ecol. Evol., 27, 244–252, https://doi.org/10.1016/j.tree.2011.11.014, 2012.
Wang, C., Duan, Q., Gong, W., Ye, A., Di, Z., and Miao, C.: An evaluation of adaptive surrogate modeling based optimization with two benchmark problems, Environ. Modell. Softw. 60, 167–179, https://doi.org/10.1016/j.envsoft.2014.05.026, 2014.
Wang, S. S.-C., Qian, Y., Leung, L. R., and Zhang, Y.: Identifying Key Drivers of Wildfires in the Contiguous US Using Machine Learning and Game Theory Interpretation, Earth's Futur., 9, e2020EF001910, https://doi.org/10.1029/2020ef001910, 2021.
Wang, S. S.-C., Qian, Y., Leung, L. R., and Zhang, Y.: Interpreting machine learning prediction of fire emissions and comparison with FireMIP process-based models, Atmos. Chem. Phys., 22, 3445–3468, https://doi.org/10.5194/acp-22-3445-2022, 2022.
Watson-Parris, D., Williams, A., Deaconu, L., and Stier, P.: Model calibration using ESEm v1.1.0 – an open, scalable Earth system emulator, Geosci. Model Dev., 14, 7659–7672, https://doi.org/10.5194/gmd-14-7659-2021, 2021.
Weng, E. S., Malyshev, S., Lichstein, J. W., Farrior, C. E., Dybzinski, R., Zhang, T., Shevliakova, E., and Pacala, S. W.: Scaling from individual trees to forests in an Earth system modeling framework using a mathematically tractable model of height-structured competition, Biogeosciences, 12, 2655–2694, https://doi.org/10.5194/bg-12-2655-2015, 2015.
Wilson, K., Goldstein, A., Falge, E., Aubinet, M., Baldocchi, D., Berbigier, P., Bernhofer, C., Ceulemans, R., Dolman, H., Field, C., Grelle, A., Ibrom, A., Law, B. E., Kowalski, A., Meyers, T., Moncrieff, J., Monson, R., Oechel, W., Tenhunen, J., Valentini, R., and Verma, S.: Energy balance closure at FLUXNET sites, Agr. Forest Meteorol., 113, 223–243, https://doi.org/10.1016/s0168-1923(02)00109-0, 2002.
Wright, I. J., Reich, P. B., Cornelissen, J. H. C., Falster, D. S., Garnier, E., Hikosaka, K., Lamont, B. B., Lee, W., Oleksyn, J., Osada, N., Poorter, H., Villar, R., Warton, D. I., and Westoby, M.: Assessing the generality of global leaf trait relationships, New Phytol., 166, 485–496, https://doi.org/10.1111/j.1469-8137.2005.01349.x, 2005.
Xu, T. and Liang, F.: Machine learning for hydrologic sciences: An introductory overview, Wiley Interdiscip. Rev. Water, 8, e1533, https://doi.org/10.1002/wat2.1533, 2021.
Zhang, J., Bras, R. L., Longo, M., and Heartsill Scalley, T.: The impact of hurricane disturbances on a tropical forest: implementing a palm plant functional type and hurricane disturbance module in ED2-HuDi V1.0, Geosci. Model Dev., 15, 5107–5126, https://doi.org/10.5194/gmd-15-5107-2022, 2022.
Zhang, Y., Ma, J., Liang, S., Li, X., and Li, M.: An Evaluation of Eight Machine Learning Regression Algorithms for Forest Aboveground Biomass Estimation from Multiple Satellite Data Products, Remote Sens.-Basel, 12, 4015, https://doi.org/10.3390/rs12244015, 2020.
Zheng, Z., Curtis, J. H., Yao, Y., Gasparik, J. T., Anantharaj, V. G., Zhao, L., West, M., and Riemer, N.: Estimating Submicron Aerosol Mixing State at the Global Scale With Machine Learning and Earth System Modeling, Earth Space Sci., 8, e2020EA001500, https://doi.org/10.1029/2020ea001500, 2021a.
Zheng, Z., West, M., Zhao, L., Ma, P.-L., Liu, X., and Riemer, N.: Quantifying the structural uncertainty of the aerosol mixing state representation in a modal model, Atmos. Chem. Phys., 21, 17727–17741, https://doi.org/10.5194/acp-21-17727-2021, 2021b.
Zheng, Z., Zhao, L., and Oleson, K. W.: Large model structural uncertainty in global projections of urban heat waves, Nat. Commun., 12, 3736, https://doi.org/10.1038/s41467-021-24113-9, 2021c.
Zhu, Q., Li, F., Riley, W. J., Xu, L., Zhao, L., Yuan, K., Wu, H., Gong, J., and Randerson, J.: Building a machine learning surrogate model for wildfire activities within a global Earth system model, Geosci. Model Dev., 15, 1899–1911, https://doi.org/10.5194/gmd-15-1899-2022, 2022.
Zuleta, D., Duque, A., Cardenas, D., Muller-Landau, H. C., and Davies, S. J.: Drought-induced mortality patterns and rapid biomass recovery in a terra firme forest in the Colombian Amazon, Ecology, 98, 2538–2546, https://doi.org/10.1002/ecy.1950, 2017.