the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
ML-SWAN-v1: a hybrid machine learning framework for the concentration prediction and discovery of transport pathways of surface water nutrients
Benya Wang
Matthew R. Hipsey
Carolyn Oldham
Nutrient data from catchments discharging to receiving waters are monitored for catchment management. However, nutrient data are often sparse in time and space and have non-linear responses to environmental factors, making it difficult to systematically analyse long- and short-term trends and undertake nutrient budgets. To address these challenges, we developed a hybrid machine learning (ML) framework that first separated baseflow and quickflow from total flow, generated data for missing nutrient species, and then utilised the pre-generated nutrient data as additional variables in a final simulation of tributary water quality. Hybrid random forest (RF) and gradient boosting machine (GBM) models were employed and their performance compared with a linear model, a multivariate weighted regression model, and stand-alone RF and GBM models that did not pre-generate nutrient data. The six models were used to predict six different nutrients discharged from two study sites in Western Australia: Ellen Brook (small and ephemeral) and the Murray River (large and perennial). Our results showed that the hybrid RF and GBM models had significantly higher accuracy and lower prediction uncertainty for almost all nutrient species across the two sites. The pre-generated nutrient and hydrological data were highlighted as the most important components of the hybrid model. The model results also indicated different hydrological transport pathways for total nitrogen (TN) export from two tributary catchments. We demonstrated that the hybrid model provides a flexible method to combine data of varied resolution and quality and is accurate for the prediction of responses of surface water nutrient concentrations to hydrologic variability.
- Article
(5183 KB) - Full-text XML
-
Supplement
(2229 KB) - BibTeX
- EndNote
Surface water nutrient concentrations have been significantly increased by human activities (Forio et al., 2015) due to urbanisation, waste discharges and agricultural intensification (Liu et al., 2012; Kaiser et al., 2013; Li et al., 2013). Increased nutrient concentrations and loads in streams alter the biogeochemical functioning and biological community structure in receiving estuaries (Jickells et al., 2014; Staehr et al., 2017), leading to an increased incidence of harmful algal blooms (Domingues et al., 2011), anoxia and hypoxia (Li et al., 2016; Testa et al., 2017) and reduced water availability (Heathwaite, 2010). Analysis of tributary water quality data over time is therefore essential to compute incoming nutrient loads, support policy and plan remediation measures.
Water quality data, however, often have constraints that make it challenging to analyse long- and short-term trends. Firstly, water quality data often have non-linear responses to environmental factors and show high-order interaction effects between different environmental variables. Moreover, nutrients can derive from different sources (point or non-point) in the landscape and are transported to receiving waters through different water pathways subject to varied catchment hydrological conditions and human intervention (Hirsch et al., 2010; Lloyd et al., 2014). Additionally, tributary nutrient datasets often are sparse in both space and time, due to the high cost of fieldwork and chemical analysis (Lamsal et al., 2006; Forio et al., 2015). Historical and current water quality monitoring programmes often use low-frequency sampling regimes on a weekly to monthly basis (Halliday et al., 2012). When monthly averaged concentrations are used, calculated nutrient loads to receiving environments such as lakes or estuaries may be poorly estimated (Cozzi and Giani, 2011), with high variability in the estimated loads (Jordan and Cassidy, 2011). It is also common to have patchy availability of nutrient species data across a study area, and combining datasets from different projects and analytical laboratories makes the analysis of long-term trends fraught with uncertainty. For instance, total nitrogen (TN) and total phosphorus (TP) concentrations within catchment outflows may have been monitored for decades, while dissolved organic nitrogen (DON) and dissolved organic carbon (DOC) concentrations may have only been monitored recently, with the increasing recognition of their ecological importance (Górniak et al., 2002; Petrone et al., 2009; Erlandsson et al., 2011). Given the hydrochemical correlation between different nutrient species and high analytical cost, there are benefits in extracting maximum information from all available nutrient data, particularly relating to changes in water quality over time (Hirsch et al., 2010). In summary, while high-quality nutrient data from tributaries are typically required as input to water quality modelling of receiving waters, the reliability and accuracy of the trend analysis of tributary data are frequently restricted by data non-linearity, limited sample size and variable nutrient availability.
Various models for constructing tributary water quality data have been developed. For example, linear models (LMs) and generalised linear models (GLMs) that use correlations between concentration (C) and flow (Q) have long played a central role in stream water quality analysis (Cohn et al., 1989; Chanat et al., 2002). Some multivariate regression models have been applied to analyse the long-term trend (Li et al., 2007; Tao et al., 2010; Greening et al., 2014) and seasonal patterns (Giblin et al., 2010; Chen et al., 2012) of surface water nutrients. For example, a weighted regression on time, discharge and season (WRTDS) was introduced by Hirsch et al. (2010) and has been applied to a number of different water quality studies (Green et al., 2014; Zhang et al., 2016a, b, c).
Meanwhile, data-driven machine learning (ML) methods are increasingly being applied to quantify relationships between soil, water and environmental landscape attributes (Lintern et al., 2018; Wang et al., 2018; Guo et al., 2019). For instance, random forest (RF), a widely used ML method, was used to model the spatial and seasonal variability of nitrate concentrations in streams (Álvarez-Cabria et al., 2016). Gradient boosting machines (GBMs) were used to quantify relationships between land-use gradients and the structure and function of stream ecology (Clapcott et al., 2012). In contrast to process-based conceptual models, ML methods simulate relationships purely from the data (Maier et al., 2014) and have the ability to incorporate different types of variables (e.g. numerical or categorised variables); this is particularly suitable for systems with complex variable interactions and non-linear response functions (Povak et al., 2014).
While both process-based and ML models can manage non-linear interactions and be used to explore long-term trends, they both have difficulty in fully extracting important hydrochemical information embedded in nutrient data. Hybrid methods have been proposed for flow forecasting, to enhance the performance of ML models by first using intermediate models to generate additional variables, which are then used for subsequent modelling. For instance, a neural network model is first applied to reconstruct surface ocean partial pressure of carbon dioxide (pCO2) climatology, which is used as an input into another neural network to predict pCO2 anomalies with other features (Denvil-Sommer et al., 2019). Similarly, Noori and Kalin (2016) used the soil and water assessment tool (SWAT) to generate baseflow and stormflow, which were then used as inputs to an artificial neural network (ANN) model to improve daily flow prediction. Both studies used hybrid models to demonstrate that pre-generated variables provided additional information that was crucial to achieving higher prediction accuracy, compared with stand-alone ANN models.
Stream flow integrates water from multiple pathways resulting in a distribution of residence times. Stream nutrients are the product of overlapping historical inputs and reaction rates, which are spatially distributed and temporally weighted within the catchment (Abbott et al., 2016). Therefore, it is beneficial to understand nutrient transport pathways from the source to receiving waters, to analyse the long- and short-term trends of stream nutrient data; this knowledge will improve management strategies to reduce nutrient transport (Tesoriero et al., 2009; Mellander et al., 2012). In the analysis of the streamflow hydrograph, separating baseflow (the long-term delayed flow from storage) and quickflow (the short-term response to a rainfall event) from total flow is a well-established strategy to better understand transport pathways (Tesoriero et al., 2009). To utilise all available nutrient data and assess the impact of different transport pathways on stream nutrient concentrations, we developed a hybrid machine learning framework for surface water nutrient concentrations (ML-SWAN) that first separated baseflow and quickflow from total flow and then built intermediate models to generate missing nutrient species within the total nutrient pool, using relationships with baseflow, quickflow, rainfall and seasonal components. The generated nutrient data were included as additional variables for a final ML prediction. RF and GBM were employed and their performance compared in stand-alone mode and as a hybrid method.
This study aimed to compare model performance for nutrient concentration prediction, to generate accurate daily nutrient data, to assess the impacts of different water transport pathways on surface water nutrient concentrations and to present a feasible framework for the application of the hybrid method for surface water nutrient prediction. It was hypothesised that the hybrid RF and hybrid GBM, which used pre-generated daily nutrient concentrations and the separated baseflow and quickflow as additional auxiliary inputs, would take advantage of the complementary strengths of hydrochemical and hydrological relationships to provide the most accurate and reliable nutrient predictions. To test this hypothesis, the hybrid RF and hybrid GBM were compared to a linear model, a multivariate weighted regression model (WRTDS), and stand-alone RF and GBM models, for the prediction of TN, TP, NH4, DOC, DON, and filterable reactive phosphorus (FRP) concentrations, at two different sites under varied hydrological conditions.
Our modelling goal in this study was to minimise the sum of the overall loss function between the predicted nutrient concentrations and measured nutrient concentrations.
where L is a loss function (e.g. squared error), yi are measured values, Xi are relevant variables, F is any approximation model, and F(Xi) or is the model-predicted value at Xi. The descriptions of different approximation models are described in the following sections.
2.1 Linear model and WRTDS model
LMs are the most commonly used tool to describe concentration–discharge (C–Q) relationships (Hirsch et al., 2010). Typically, a log transformation is often applied to C and Q data (Crowder et al., 2007; Meybeck and Moatar, 2012; Herndon et al., 2015), with the linear model then described as
where C is nutrient concentration and Q is total flow. In this study, the linear model was used as a benchmark for other models. The fitted slope β0 can represent the base nutrient concentration in a stream, while β1 can describe relationships between hydrological and biogeochemical data. The WRTDS model was also used (Hirsch et al., 2010) and can be described as
where JD is the Julian day and ε is unexplained variation. β2JD is used to represent the long-term trend from year to year, while β3cos (JD) and β4sin (JD) are used to describe the seasonal variation in stream nutrient concentrations. To calculate the Julian Day for use in Eq. (3), the days since 1 January 1970 were first calculated and then multiplied by 2π. WRTDS advances the simpler linear model in two aspects. Firstly, the additional components in the equation allow a consideration of seasonal and long-term patterns and make the WRTDS model more able to describe stream nutrient concentrations across the year. Secondly, unlike the linear model, whose parameters are constant in time, WRTDS adjusts the parameters in a gradual manner throughout Q, JD space. This is accomplished by applying a weighted regression for the estimation of log (C), where the weights on each observation are based on three distances between the observation (Qo, JDo) and the estimation point (Qi, JDi), which are (1) the time distance between JDo and JDi, (2) the seasonal distance between the time of year at JDo and the time of year at JDi, and (3) the discharge distance between log (Qo) and log (Qi) (Hirsch et al., 2010; Green et al., 2014). Thus, log (C) is considered to be locally linearly related to log (Q), JD, sin (JD) and cos (JD).
2.2 Random forest and gradient boosting machines
RF and GBMs are ensemble models that combine multiple base learners inside the model to improve the prediction performance (Ishwaran and Kogalur, 2010; Singh et al., 2014). The ensemble methods are the main difference between RF and GBM. In RF, bootstrap aggregating is used to resample the original dataset with replacement. Hence, datasets with partial data are generated and then used to build individual base learners. Unlike bootstrap aggregating, GBM iteratively generates a sequence of base learners, where each successive base learner is built for the residual prediction of the preceding base learner (Friedman, 2001, 2002). The probability with which data points are selected for the next training set is not constant and equal for all data points. The selection probability increases for data points that have been misestimated in the previous iteration; data points that are difficult to classify would receive higher selection probabilities than easily classified data points (Yang et al., 2010; Erdal and Karakurt, 2013).
For RF and GBM, the most commonly used base learner is a classification and regression tree (CART). A CART model is built to split the dataset into different nodes (Breiman et al., 1984): and for numeric variables or and for categorised variables, where i and j are the sample indices, a is a numerical variable, v is one of the values of a variable, d is a categorised variable, and c is one of the values of d variable. To split the dataset at a or d, the sum of least-square error of the two nodes is calculated for a regression task as
where yl and yr are observations in two split nodes and and are the average y in that node. The split is chosen among all candidate variables and values to minimise this error. This splitting process is applied from the root to the terminal node, which creates a tree structure for the model (Erdal and Karakurt, 2013). A CART can be used both for classification and regression problems due to this tree structure (Coops et al., 2011). However, a single CART can sometimes oversimplify variable interactions and may lead to low prediction performance (McBratney et al., 2000; Cutler et al., 2007; Coopersmith et al., 2010). This drawback can be overcome by the ensemble method that generates many resampled datasets and creates various CARTs to achieve higher accuracy (Breiman, 2001) and more stable results when facing slight variations in input data (Martínez-Rojas et al., 2015). New data input is thus evaluated against all trees created in the ensemble model, and each tree votes using the main class or the averaged values in the terminal node. The class with the maximum votes will be used for a classification model, and the averaged predicted value from all trees is used for a regression model (Singh et al., 2014; Belgiu and Drăgu, 2016). It is found that ensemble methods in RF and GBM can significantly improve the prediction accuracy of CART (Ismail and Mutanga, 2010; Erdal and Karakurt, 2013).
Compared to LM and WRTDS models, one drawback of RF and GBM, as well as many ML methods in general, is that there is no specific equation in GBM or RF to directly demonstrate model structures. However, GBM and RF do provide the relative importance of each variable, which is based on the empirical improvement in the loss function due to the split on the specific variable in a tree (Povak et al., 2014; Puissant et al., 2014). The improvement of a certain variable was averaged over all trees and used as the relative importance of that variable for the final model. This relative importance serves as the key index to understand the model structure of RF and GBM (Makler-Pick et al., 2011).
2.3 Baseflow separation
Total flow is commonly conceptualised as including baseflow and quickflow components (Meshgi et al., 2015). Baseflow separation techniques use the time-series record of streamflow to extract the baseflow and quickflow signatures from the total flow. This can be done by using graphical methods to identify the intersection between baseflow and the rising and falling limbs of the quickflow response (Szilagyi and Parlange, 1998) or by filtered methods which process the entire stream hydrograph to derive a baseflow hydrograph (Furey and Gupta, 2001). In this study, the three-pass filtered method was applied for baseflow separation; the quickflow was first estimated as described below (Lyne and Hollick, 1979; Nathan and McMahon, 1990), and then baseflow was calculated:
where QFi is the filtered quickflow for the ith sampling instant, QFi−1 is the filtered quickflow for the previous sampling instant to i and α is the filter parameter with a value of 0.925 for daily flow as recommended by Nathan and McMahon (1990). Baseflow is then calculated as .
2.4 Performance evaluation metrics
In this study, the root mean squared error (RMSE) and the Nash–Sutcliffe model efficiency coefficient (MEF) were used to compare model performance. The RMSE is a measure of overall error between the predicted and measured data and returns an error value with the same units as the data, which is given by the following equation:
where n is the number of data samples. RMSE varies from 0 to +∞, and a perfect model would have RMSE of 0. The MEF is a dimensionless “goodness-of-fit” measure which can vary from −∞ to 1, with a value of 1 indicating a perfect fit and 0 indicating that the mean of the measured values performs as well as the model. The MEF can be calculated as
where is the mean of the measured values. Note that the predicted and measured nutrient values were normalised to [0, 1] in this study to compare model performance across different nutrient species.
2.5 Overview of modelling processes
The main aims of this research is to test the hybrid model, rebuild the historical nutrient data, and explore the short- and long-term nutrient changes. The first step is verifying the model performance. In this case, the data were randomly divided into 80:20. Different models were built and tuned on the training dataset (80 %) and tested on the testing dataset (20 %). To further test model uncertainty and stability, the divided and tested processes were repeated 30 times except for WRTDS. After this, all data points including the testing data were then used to rebuild the historical nutrient data. Five-fold cross validation (CV) was done on the training dataset to tune the model parameters. Leave-one-out cross validation (LOOCV) was used in WRTDS to predict daily nutrient concentrations; LOOCV is the default cross-validation method in the EGRET (Exploration and Graphics for RivEr Trends) package. In that method, one data point was excluded at a time from the whole dataset, all other data points were used to build the model, and the excluded point was used for testing the model performance. This process was repeated for all data points. The performance of all six methods (LM, WRTDS, RF, GBM, hybrid RF and hybrid GBM) was evaluated on the testing dataset. WRTDS was run through the EGRET package (Hirsch and De Cicco, 2015) in R to produce daily concentrations for six nutrient species (TP, TN, DON, DOC, NH4 and FRP). The default settings specified by the user guide (Hirsch and De Cicco, 2015) were used. RF and GBM models were built through the H2O package in R.
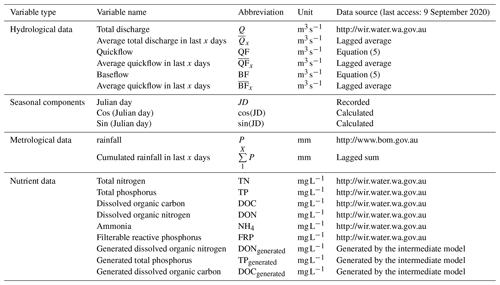
The overall processes of ML-SWAN can be divided into three stages (Fig. 1). The first stage was baseflow separation using the EcoHydRology package (Fuka et al., 2018). The generated baseflow, quickflow, total flow and rainfall were further transformed into lagged data (the averaged values over the previous 3, 7 and 15 days) to capture any short-term impacts of different water pathways and rainfall on stream nutrients. JD, cos (JD) and sin (JD) were also calculated for RF and GBM to include seasonal and long-term impacts. A description of all the variables used is given in Table 1.
The second stage of ML-SWAN was to build intermediate RF and GBM models that generated daily nutrient concentrations. For the intermediate RF and GBM models, only lagged hydrological data (including total flow, baseflow and quickflow), lagged rainfall and seasonal components on the training dataset were used. Nutrients were not used as a predictor in the intermediate model. Note that, in this study TP, TN, DOC and DON were selected to be generated in the second step. If one nutrient was considered as the final target, the other three nutrients were used to generate daily data. For instance, daily TP, DOC and DON were generated as additional variables to predict TN. In that case, the missing TP, DOC and DON were generated by the intermediate model for the training dataset and the testing dataset. Daily TN, TP, DOC and DON data were generated and used for the final predictions. These nutrients were selected since they may be generated from similar sources or are important components of the total nutrient load. For instance, DOC and DON may both be generated from dissolved organic matter (DOM) (Seitzinger et al., 2002; Bernal et al., 2005; Filep and Rékási, 2011). In the catchments studied here, DON can be a dominant component of TN (Nice et al., 2009; Petrone, 2010; Bourke et al., 2015). The selection of DOC and DON for pre-generation may not necessarily be appropriate for other catchments. The selection of nutrients for pre-generation depends on data availability in the dataset. The use of different species of the same nutrients (N or P) can generally improve model performance.
The third stage of ML-SWAN built an additional hybrid model using the training data, which has generated nutrient data by the intermediate models, lagged hydrological data, lagged rainfall data and seasonal components. Note that at this stage, the only difference between stand-alone ML and hybrid ML methods was that stand-alone ML did not use pre-generated daily nutrient data.
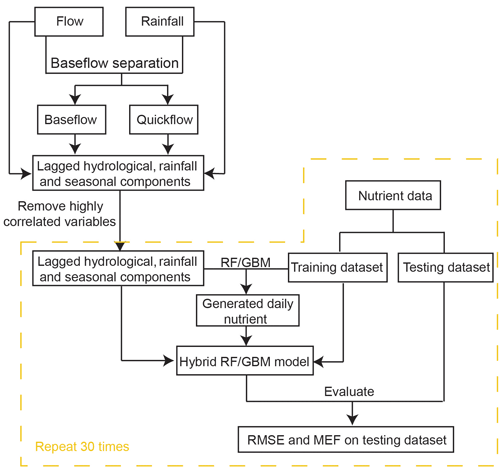
To test the generalisability of the hybrid framework, two sites in Western Australia (Ellen Brook and Murray River) were selected as study areas. Ellen Brook and Murray River are key tributaries for the Swan–Canning Estuary and Peel–Harvey Estuary (Fig. 2), respectively, and have different hydrological conditions. The Swan–Canning Estuary is located adjacent to the Perth metropolitan area, with an area of approximately 40 km2. The catchment comprises 30 catchments, which drain approximately 2090 km2 (Kelsey et al., 2010). Ellen Brook is the largest sub-catchment in the Swan–Canning catchment, comprising 34 % (716 km2) of the total catchment area. Ellen Brook is an ephemeral river with no flow recorded during summer and the early autumn months (Table 2). The dominant land use in Ellen Brook is agricultural and grazing land. Ellen Brook is one of the highest contributors of TN and TP to the Swan–Canning Estuary (Swan River Trust, 2009). Bassendean sands and duplex Yanga (sand over clay) soils dominate the Ellen Brook catchment. Bassendean sands have very low phosphorus retention indices (PRIs), while Yanga soils have low PRIs in their upper horizon and become waterlogged in winter, promoting the release of retained nutrients to the stream (Kelsey et al., 2010).
The Peel–Harvey Estuary is located approximately 75 km south of the Swan–Canning Estuary, and the Serpentine, Murray and Harvey Rivers drain into the estuary (Fig. 2). The total catchment area of the estuary is approximately 11 930 km2. The Murray River catchment is dominated by deep grey sands, loams clay and peats (Ruibal-Conti et al., 2013), agricultural land use, and natural reserves, and it contributes about 40 % of annual TN loads and 7 % of annual TP loads to the estuary (Kelsey et al., 2011).
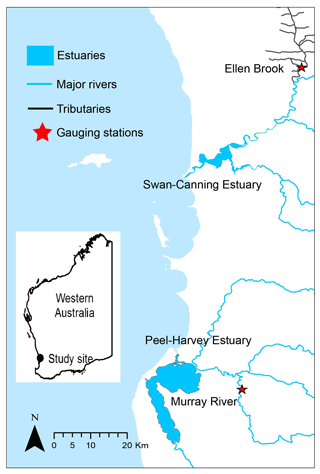
Both Swan–Canning Estuary and Peel–Harvey Estuary experience a Mediterranean climate with cool, wet winters (June–August) and hot, dry summers (December–March). The long-term average annual rainfall varies from 1300 mm on the coast to 800 mm in the south-east of the catchment area (1975–2009, Bureau of Meteorology station), and about 90 % of the rain falls between April and October. Sample size and the first measurement year of six nutrients species are listed for the two study sites in Table 3. TN, TP, NH4 and FRP have been monitored for decades, while DOC and DON have only been measured in recent years, with limited sample size. Several historical nutrient datasets were combined but significant changes occurred in water sampling devices and analytical instrumentation over the past decades. These changes can increase the complexity of nutrient data. For instance, auto-samplers sampled any time regardless of weather conditions (e.g. during the rainfall), while grab samples were typically collected under fine weather conditions due to safety concerns.
4.1 Comparison of prediction accuracy between six methods
Overall, the scaled RMSE reduced from LM, WRTDS, stand-alone ML and hybrid ML for all nutrients except NH4, and the same pattern was found for MEF in both Ellen Brook and Murray River (Fig. 3). The linear model had the worst performance: the scaled RMSE was significantly higher and MEF was significantly lower than the other models, for all six nutrients and across both sites. WRTDS generally had higher RMSE and lower MEF than the stand-alone ML, although it achieved similar results to stand-alone ML for FRP and NH4 at both sites. LOOCV was used in WRTDS, and only one set of results was generated, compared to 30 RMSE and MEF values for other methods. This results in a shortened line for WRTDS in Fig. 3, instead of the interquartile ranges (IQR=75th percentile − 25th percentile) presented for the other methods. LOOCV can sometimes overestimate the model performance as only one sample was tested at a time; in contrast, 20 % of the independent testing data were tested in the other five models. LOOCV can also have a higher variance than other CV methods (Li, 2016). As such, the WRTDS results are not directly comparable to the other methods.
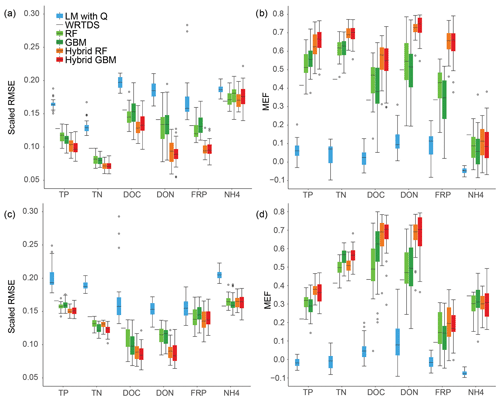
Figure 3Model performance across six nutrients and the two sites: (a) RMSE and (b) MEF for Ellen Brook; (c) RMSE and (d) MEF results for Murray River.
Stand-alone ML achieved results that placed it between WRTDS and hybrid ML. Stand-alone GBM achieved the highest accuracy for NH4 prediction in Murray River. Hybrid RF and hybrid GBM had the lowest RSME and highest MEF for all nutrients except NH4, in Ellen Brook and Murray River (Fig. 3). Compared to the stand-alone ML, the hybrid ML also had much lower prediction uncertainty, in that the RMSE and MEF had narrower IQR than that of the stand-alone ML, especially for DON and FRP prediction in Ellen Brook and DOC prediction in Murray River. The use of pre-generated daily nutrient data was the only difference between hybrid ML and stand-alone ML. This means that the generated nutrients provided additional information for the hybrid model that allowed more stable results. Interestingly, while the hybrid ML had better performance than the stand-alone ML, there was no significant difference in performance between the hybrid RF and hybrid GBM, though they showed differences between different nutrient species. For instance, hybrid RF achieved slightly better performance for DOC in Ellen Brook, while hybrid GBM had lower RMSE for DOC in Murray River. There was no significant performance difference between stand-alone RF and GBM.
In summary, the hybrid ML had the best performance amongst the six methods, followed by stand-alone RF and GBM. WRTDS was better than the linear model but could only achieve results similar to stand-alone RF and GBM for NH4 prediction in Ellen Brook and for NH4 and FRP prediction in Murray River.
4.2 Generated daily TN in Ellen Brook
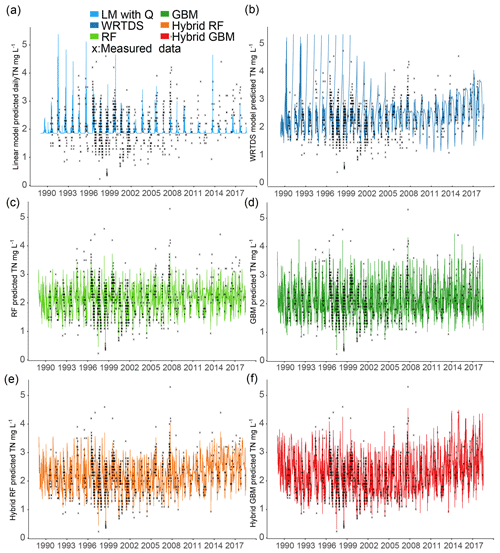
Model performance for six nutrients was compared in the last section. To make this section more concise, these six models were then compared in their ability to generate daily TN in Ellen Brook from 1 January 1989 to 16 July 2018 (Fig. 4). The daily TN in Murray River and daily TP in both sites were also generated (see results in the Supplement). TN was selected because TN is the most important and most frequently measured nutrient in many places. This hybrid method can also be used for other nutrients. Note that all data points (not just the 80 % training dataset) were used to generate daily TN.
The LM performed very poorly for TN prediction; low-concentration samples () were all underestimated, and some extremely high concentrations were incorrectly generated due to the high flow (Fig. 4a). There were some seasonal patterns in the generated TN which come from the flow data. LM only used total flow to predict nutrient concentrations, while other important hydrological processes were ignored. Thus the oversimplified LM had high errors in nutrient prediction (Fig. 4), and this method might be more suitable for solutes that are not substantially bioactive (e.g. SiO2, Ca2+, Mg2+, Cl−) (Stallard and Murphy, 2014). The WRTDS captured some seasonal patterns of TN (from 2008 to 2018) but still had problems predicting TN between 1989 and 1996; some extremely high values were generated, and were overestimated. Some high values (e.g. TN in 2008) were underestimated (Fig. 4b). Stand-alone ML and hybrid ML generated similar daily TN data but varied in the detail. These models successfully captured the low-concentration data and the seasonal pattern of TN. Unlike results by WRTDS, the generated TN by stand-alone ML and hybrid ML have a more consistent seasonal pattern from 1989 to 2018. The RF and hybrid RF both underestimated a few high-concentration data (), compared to GBM and hybrid GBM, although hybrid RF still showed better performance than RF. For instance, high-concentration data in 2007 and again from 2014 to 2017 were successfully predicted by hybrid RF but underestimated by RF. Compared to stand-alone GBM, the hybrid GBM achieved lower errors for high-concentration data.
Apart from the better performance for high-concentration data, another difference between stand-alone ML and hybrid ML was that the long-term trend in TN was consistent in stand-alone ML, but this trend fluctuated in hybrid ML. For instance, hybrid GBM results fluctuated from 1989 to 1999 and then showed an increasing long-term trend from 2005 to 2018, in addition to the seasonal fluctuation. The pre-generated nutrient is the only difference between stand-alone model and hybrid model. If there are long-term trends in nutrient concentrations (e.g. TN), similar trends should also exist in the components of TN (either DON or dissolved inorganic nitrogen). The pre-generated nutrients emphasise this impact on the hybrid model. This suggests that the generated nutrient data could provide additional information that allowed the hybrid ML to capture long-term trends; this information was not included in the seasonal components but existed in the generated nutrient data.
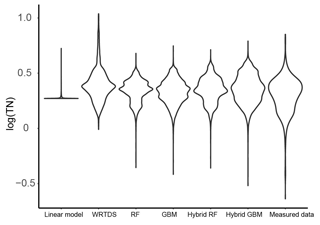
Figure 5The distribution of the daily TN generated by the six models and that of the measured TN data in Ellen Brook.
The distribution of the TN data generated by the six models was compared to the distribution of the measured TN data (Fig. 5). Similar to the results shown in Fig. 4, hybrid GBM had the most similar distribution to the measured TN data. Only a few low- and high-concentration data were incorrectly predicted by the hybrid GBM. Hybrid RF also achieved a distribution similar to the measured data, but more extreme-value data were underestimated compared to the hybrid GBM. Stand-alone GBM and RF showed a similar distribution to the hybrid GBM and RF with less accuracy in the extreme data. Overall, GBM (either stand-alone model or hybrid model) could have a better distribution than RF. WRTDS generated some extremely high data and underestimated many low-concentration data, which is also seen in Fig. 4b. The linear model incorrectly predicted most of the TN data. The results in both Figs. 4 and 5 showed that hybrid GBM achieved the best simulated daily TN data, followed by hybrid RF, stand-alone GBM and RF. WRTDS and LM generated large biases in TN prediction.
The hybrid ML models predicted most of the extreme concentrations (Figs. 4 and 5), and only a few points were under-predicted. The limited number of extreme data and the model structure that tried to balance the overall trend prediction with extreme data prediction can cause under-prediction. For example, higher weights can be set up for extreme data during the model training process to force model to over-predict the value for extreme concentrations, which may reduce the accuracy for overall trend prediction. In this study, our target is to understand the long-term nutrient trend. Therefore, we did not use this technique during the model training process.
4.3 Comparison of variable importance in hybrid GBM for TN prediction
The daily data generated by the hybrid GBM showed a lower RMSE and better distribution than stand-alone ML, WRTDS and LM (Figs. 4 and 5). Compared to LM, WRTDS and simple CART models, one drawback of RF and GBM, as well as many ML methods in general, is that there is no specific equation in GBM or RF to directly demonstrate model structures. However, GBM and RF do provide the relative importance of each variable, which is based on the empirical improvement in the loss function due to the split on the specific variable in a tree (Povak et al., 2014; Puissant et al., 2014). The improvement of a certain variable was averaged over all trees as the relative importance for the final model. This relative importance serves as the key index to understanding the model structure of RF and GBM (Makler-Pick et al., 2011).
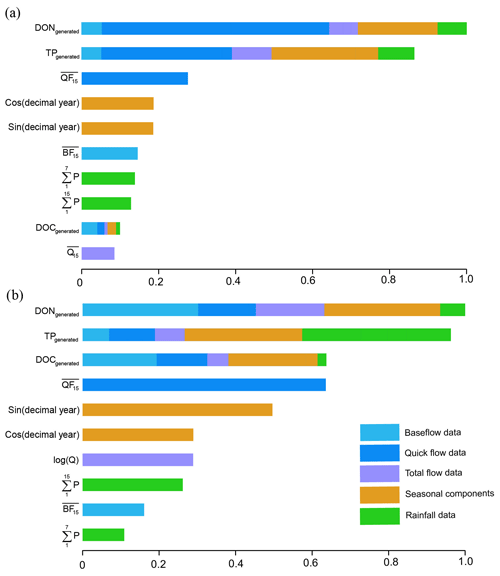
Figure 6Variable importance in the hybrid GBM for TN prediction in (a) Ellen Brook and (b) Murray River.
The variable importance for TN prediction by hybrid GBM in Ellen Brook and Murray River is presented in Fig. 6. The variable importance in the intermediate models is also included, and the length of coloured sections represents the importance of those variables in the hybrid GBM or intermediate GBM. The importance was scaled according to the most important variable. The generated DON and TP ranked as the first two critical variables in Ellen Brook, while all three generated nutrients were listed as the most important variables in Murray River. This suggests that the generated nutrients do provide critical information to the model and improve model performance. The quickflow was most important for the generated DON and TP, as well as the TN itself in Ellen Brook. The impacts of quickflow decreased, and baseflow, seasonal components and rainfall data become more important for TN prediction in Murray River. This difference in variable importance reflects different catchment characteristics across the two sites and therefore different hydrological and hydrochemical processes controlling TN concentrations. The total flow was not of high importance at either site, which suggests that baseflow or quickflow had more impact on surface water TN. Moreover, TN concentrations were affected by more variables in Murray River than in Ellen Brook.
5.1 Different sources of TN in Ellen Brook and Murray River
Hydrological conditions, specific sub-catchment characteristics and the chemical properties of nutrients can all impact surface water nutrient concentrations (Barron et al., 2009; Moatar et al., 2016), nutrient partitioning (Ruibal-Conti et al., 2013) and nutrient transport (Burt and Pinay, 2005; Tesoriero et al., 2009). TN prediction in Murray River was impacted by more variables than in Ellen Brook (Fig. 6), suggesting more complex relationships in Murray River.
Quick flow is composed of runoff, interflow and direct precipitation (Brodie and Hostetler, 2005) and was shown to be important for TN prediction in Ellen Brook. Direct precipitation, however, did not have a large impact on TN (the green bars in Fig. 6); this suggests that runoff and interflow were important for TN concentrations. Baseflow can account for (on average) 53 % of annual stream discharge in Ellen Brook, but baseflow was not of high importance for TN prediction in this study. This may occur due to low TN concentrations in the baseflow (Barron et al., 2009), large areas of low nutrient-retaining sandy soils in the Ellen Brook catchment, and high nutrient transport efficiency in quickflow and first flush. Mellander et al. (2012) quantified nutrient transport pathways in agricultural catchments and found that quickflow was only 2 %–8 % of total flow, but it can transport up to 50 % of TP. Gunaratne et al. (2017) found that the seasonal first flush was only 30 % of runoff volume but contained 40 %–70 % of the nutrient load.
Note that the median TN in Ellen Brook (2.1 mg L−1) is significantly higher than that in Murray River (0.67 mg L−1) which can be explained to some extent by the large area of grazing lands in Ellen Brook. Previous investigations in south-eastern Australia (Adams et al., 2014), New Zealand (Davies-Colley et al., 2004) and north-western Europe (Conroy et al., 2016) all suggested that livestock can increase TN discharge to the receiving water bodies. Most of the piggeries and poultry farms in the Swan–Canning catchment are located in Ellen Brook catchment (Kelsey et al., 2010), which has the highest TN and TP discharge loads. Thus the large grazing areas, piggeries and poultry farms and low nutrient-retaining sandy soils may explain the importance of quickflow for TN prediction and high TN concentrations in Ellen Brook.
Baseflow is derived from groundwater discharge to streams and the slow drainage of water stored in local wetlands (Kelsey et al., 2010). Baseflow is highlighted as an important variable for TN prediction in Murray River. The Murray River catchment has large areas with high nutrient-retaining soils (high PRI) (Kelsey et al., 2011) and relatively low TN concentrations, and it is likely that groundwater makes significant contributions to TN in Murray River. Ruibal-Conti et al. (2013) previously found that variability in TN is strongly associated with variability in flows in Murray River. Our results extend this finding, in that both baseflow and quickflow likely impact TN in the river.
It is noted that seasonal components including sin (JD), and cos (JD) showed significantly higher importance in Murray River. This may because seasonal information is captured in other inputs in Ellen Brook (e.g. quickflow and baseflow). But the main reason is the stronger seasonal TN signals in Murray River compared to Ellen Brook. This finding is supported by the generated daily TN data for Murray River (see results in Supplement S2). Natural reserves occupy large areas of the Murray River catchment, and this may increase seasonal signals. Additionally, the lagged quickflow, baseflow and rainfall were generated (for the previous 3, 7 and 15 days), but only the lagged 15 d baseflow and quickflow were ranked as important variables for both Ellen Brook and Murray River. This suggests a timescale of nutrient transport in the sub-catchments and likely reflects soil permeability and geology; long hydrochemical recessions from storm events may prolong their impact on the ecological status of receiving rivers (Mellander et al., 2012).
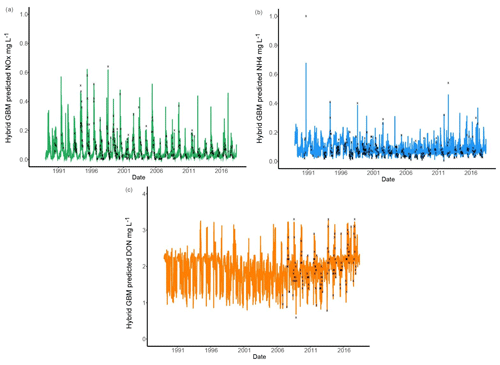
Six models were compared for nutrient predictions and the hybrid GBM model achieved the highest accuracy (Figs. 3 and 5). The long-term changes in TN have been discussed in previous sections. To understand the long-term changes in other nitrogen species across the year, the hybrid GBM was then applied to generate daily DON, NH4 and NOx in Ellen Brook from 1 January 1989 to 16 July 2018 (Fig. 7). The generated DON has much higher concentration than NH4 and NOx. This is consistent with previous investigations in this study area that DON was the dominant form of TN in both surface water and groundwater (Nice et al., 2009; Petrone, 2010; Bourke et al., 2015). There is no clear long-term patterns in generated NH4 and NOx; however, an increasing long-term trend in generated DON can be found from 2006 to 2018. There is also an increasing trend in TN from 2005 (Fig. 4), suggesting DON was the main reason for the increasing TN concentrations. DON is often assumed to be relatively slow to react, but depending on the source of DON, it can turnover rapidly, thereby constituting an active contributor to the eutrophication of surface waters (Petrone et al., 2009).
5.2 Can we improve our understanding of historical nutrient conditions using a contemporary data?
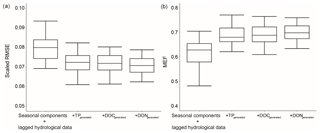
The generated nutrient data provided additional information to enhance the hybrid model performance (Figs. 3 and 5). To assess the individual impact of a generated nutrient, we did a simple test that sequentially added generated TP, DOC and DON data to the base GBM (only seasonal components and lagged hydrological data) and evaluated RMSE and MEF for TN prediction. This process was repeated 30 times and the results are presented in Fig. 8.
The RMSE significantly decreased when generated TP was added as an additional variable. DOC and DON only have 297 and 129 data, respectively, and were only measured in recent years, while TP has more than 1000 data and has been measured since 1990 (Table 3). However, DOC and DON could still improve model performance (Fig. 8), and the generated DON was ranked as the most important variable across both sites (Fig. 6). The medium RMSE slightly decreased when both generated DOC and DON were added. Moreover, the generated DOC and DON also reduced the model uncertainty, such that the IQRs became narrower than model results without the generated nutrients.
Our results suggest that the recent DON and DOC data improved understanding of historical TN. It is not uncommon to have a similar data structure when several datasets are combined or new measurements are added to a project. While there were no DON data prior to 2006 in Ellen Brook, daily DON can be generated back to 1990 with the help of generated TN, DOC and TP data; DON had the highest MEF among the six nutrients (Fig. 3). This hybrid method provides a feasible process to fully utilise all available nutrient data to accurately fill gaps in either historical or recent nutrient datasets.
5.3 A comprehensive comparison of six models
Monitoring, modelling and forecasting water quality inputs are essential to support the management of the quality of receiving waters while responding to current anthropogenic stressors (Holguin-Gonzalez et al., 2013; Schnoor, 2014). The performances of six models were comprehensively compared, in an exploration of historical and contemporary nutrient data across two study sites. LM had the highest error while stand-alone RF and GBM had similar error. This agrees with previous findings by Erdal and Karakurt (2013) that RF and GBM models achieved similar correlation coefficients (R) for streamflow forecasting. Ismail and Mutanga (2010) also reported that RF and GBM increased the R of a single CART by 10.01 % and 9.59 %, respectively.
The performance of WRTDS, as well as many conceptual models, is often reliant on a prescribed set of input information, which can account for variance in nutrient concentrations but may miss some important processes for certain rivers (e.g. baseflow in this study). This can compromise the performance of WRTDS for nutrient prediction. Moreover, hydrological and chemical processes within the systems are typically ignored by many conceptual models, which may exclude important hydrochemical information. By contrast, some complex conceptual models may include these hydrochemical processes but are often constrained by insufficient nutrient data to calibrate and validate the models. Some simplifications may be made to account for lack of data, but the simplifications may often weaken model performance. The hybrid framework presented in this study has overcome the challenge caused by data paucity by building intermediate models to generate missing nutrient data and then using this additional hydrochemical information to improve final model performance.
The hybrid models developed in this study were able to take advantage of the complementary strengths of both hydrochemical (additionally generated nutrient data) and hydrological (lagged data) information. This was particularly the case for the prediction of high nutrient concentrations, where the hybrid models were shown to outperform the stand-alone RF and GBM, in terms of accuracy, reliability and value distribution. Improved accuracy in the hybrid model was achieved by using intermediate models, although these intermediate models may also have a relatively high error (similar to stand-alone RF and GBM). However, if the improved model performance is higher than the introduced error, the results are manageable. Similar results were also found in Hunter et al. (2018), who compared a hybrid process-driven and ANN model with the stand-alone ANN model and the process-driven model. In their study, the hybrid also achieved the best performance followed by stand-alone ANN. The process-driven benchmark model had a significantly lower accuracy than the other two models.
A limitation of the hybrid modelling approach, however, is that it requires the time and expertise to develop intermediate models for generating additional nutrient data. Prior knowledge also plays an important role in identifying the variables for pre-generation. Some statistical methods (e.g. the correlation test, simple linear model) can be helpful to identify these variables if there is no clear theoretical or conceptual understanding on which to base the selection of the important variables.
In this study, we tested the generalised performance of the hybrid model across six nutrient species and two tributaries. We also note that nutrients may not always be the critical variables targeted for pre-generation; the pre-generated DOC was ranked as having low importance for Ellen Brook and produced only a slight improvement in the performance of the hybrid model for NH4.
5.4 The application of ML methods for hydrological modelling
There were constraints in the nutrient datasets in this research, and similar constraints commonly exist in other study areas. Many nutrient datasets contain important information, but sometimes it can be challenging to directly combine or utilise them. ML methods provide a feasible approach to preprocess these datasets or combine them. In this study, the concentrations of missing nutrient species were first predicted by the intermediate ML method and then used as inputs for another ML method for final predictions. The pre-generation of missing data and pre-modelling hydrological analysis were critical components of the hybrid model and allowed the identification of the impact of different hydrological transport pathways for TN export from the two tributary catchments. The hybrid ML methods were further applied to generate nutrient data for eight tributaries, and the generated data have since been used as inputs to an estuary prediction model, which simulates and forecasts nutrient concentrations in the previous and next 5 d in the Swan–Canning Estuary (Huang et al., 2019). The modelling methods and strategies developed in the work presented here can be easily applied to other study areas. Overall, ML methods provide a flexible and feasible solution to explore the underlying relationships, reconstruct spatial and temporal datasets, and combine different models.
A hybrid machine learning model was developed, and its performance tested on six nutrients and two estuary tributaries and compared with alternative modelling approaches. The hybrid ML model exhibited higher prediction accuracy and lower prediction uncertainty than stand-alone ML, WRTDS and LM for almost all nutrients. The pre-generation of missing data and pre-modelling hydrological analysis were critical components of the hybrid model and allowed the identification of the impact of different hydrological transport pathways for TN export from the two tributary catchments. The results of this study demonstrate the advantages of using hybrid models for high temporal resolution nutrient prediction; the results also demonstrate the use of the hybrid model for re-analysis of historical data in the light of contemporary data. Modelling strategies for different modelling targets and dataset structures have also been discussed. The modelling framework presented here can aid others to fully use all available nutrient data to generate accurate nutrient predictions.
The data and the data sources used in this study are cited and explained in the text. The current version of model is available from the project website: https://github.com/benyawang-uwa/daily-nutrient-prediction (last access: 9 September 2020) under the MIT licence. The exact version of the model used to produce the results used in this paper is archived on Zenodo (https://doi.org/10.5281/zenodo.3739611, Wang, 2020).
The supplement related to this article is available online at: https://doi.org/10.5194/gmd-13-4253-2020-supplement.
BW, MRH and CO contributed to the development of the methodology and designed the experiments, and BW carried them out. BW developed the model code and performed the simulations. BW prepared the paper with contributions from all coauthors.
The authors declare that they have no conflict of interest.
The authors acknowledge Peisheng Huang and Brendan Busch for providing the historical nutrient data.
Benya Wang was supported by a postgraduate scholarship provided by the CRC for Water Sensitive Cities. Matthew R. Hipsey received funding support from the Australian Research Council (project LP150100451).
This paper was edited by Thomas Poulet and reviewed by Thu Huong Thi Hoang and one anonymous referee.
Abbott, B. W., Baranov, V., Mendoza-Lera, C., Nikolakopoulou, M., Harjung, A., Kolbe, T., Balasubramanian, M. N., Vaessen, T. N., Ciocca, F., Campeau, A., Wallin, M. B., Romeijn, P., Antonelli, M., Gonçalves, J., Datry, T., Laverman, A. M., de Dreuzy, J. R., Hannah, D. M., Krause, S., Oldham, C., and Pinay, G.: Using multi-tracer inference to move beyond single-catchment ecohydrology, Earth-Science Rev., 160, 19–42, https://doi.org/10.1016/j.earscirev.2016.06.014, 2016.
Adams, R., Arafat, Y., Eate, V., Grace, M. R., Saffarpour, S., Weatherley, A. J., and Western, A. W.: A catchment study of sources and sinks of nutrients and sediments in south-east Australia, J. Hydrol., 515, 166–179, https://doi.org/10.1016/j.jhydrol.2014.04.034, 2014.
Álvarez-Cabria, M., Barquín, J., and Peñas, F. J.: Modelling the spatial and seasonal variability of water quality for entire river networks: Relationships with natural and anthropogenic factors, Sci. Total Environ., 545–546, 152–162, https://doi.org/10.1016/j.scitotenv.2015.12.109, 2016.
Barron, O., Donn, M., Furby, S., Chia, J., and Johnstone, C.: Groundwater contribution to nutrient export from the Ellen Brook catchment, available at: http://www.clw.csiro.au/publications/waterforahealthycountry/2009/wfhc-groundwater-Ellen-Brook-catchment.pdf (last access: 9 September 2020), 2009.
Belgiu, M. and Drăgu, L.: Random forest in remote sensing: A review of applications and future directions, ISPRS J. Photogramm. Remote Sens., 114, 24–31, https://doi.org/10.1016/j.isprsjprs.2016.01.011, 2016.
Bernal, S., Butturini, A., and Sabater, F.: Seasonal variations of dissolved nitrogen and DOC : DON ratios in an intermittent Mediterranean stream, Biogeochemistry, 75, 351–372, https://doi.org/10.1007/s10533-005-1246-7, 2005.
Bourke, S., Hammond, M., and Clohessy, S.: Perth Shallow Groundwater Systems Investigation: North Lake, available at: https://www.water.wa.gov.au/__data/assets/pdf_file/0016/7432/108960.pdf (last access: 9 September 2020), 2015.
Breiman, L.: Random forests, Mach. Learn., 45, 5–32, 2001.
Breiman, L., Friedman, J., Stone, C. J., and Olshen, R. A.: Classification and regression trees, CRC Press, Boca Raton, 1984.
Brodie, R. and Hostetler, S.: A review of techniques for analysing baseflow from stream hydrographs, in: Proceedings of the NZHS-IAHNZSSS 2005 Conference, Auckland, New Zealand, 2005.
Burt, T. P. and Pinay, G.: Linking hydrology and biogeochemistry, Prog. Phys. Geogr., 3, 297–316, https://doi.org/10.1067/mva.2002.123763, 2005.
Chanat, J. G., Rice, K. C., and Hornberger, G. M.: Consistency of patterns in concentration-discharge plots, Water Resour. Res., 38, 10–22, https://doi.org/10.1029/2001WR000971, 2002.
Chen, Y., Liu, R., Sun, C., Zhang, P., Feng, C., and Shen, Z.: Spatial and temporal variations in nitrogen and phosphorous nutrients in the Yangtze River Estuary, Mar. Pollut. Bull., 64, 2083–2089, https://doi.org/10.1016/j.marpolbul.2012.07.020, 2012.
Clapcott, J. E., Collier, K. J., Death, R. G., Goodwin, E. O., Harding, J. S., Kelly, D., Leathwick, J. R., and Young, R. G.: Quantifying relationships between land-use gradients and structural and functional indicators of stream ecological integrity, Freshw. Biol., 57, 74–90, https://doi.org/10.1111/j.1365-2427.2011.02696.x, 2012.
Cohn, T. A., Delong, L. L., Gilroy, E. J., Hirsch, R. M., and Wells, D. K.: Estimating constituent loads, Water Resour. Res., 25, 937–942, https://doi.org/10.1029/WR025i005p00937, 1989.
Conroy, E., Turner, J. N., Rymszewicz, A., O'Sullivan, J. J., Bruen, M., Lawler, D., Lally, H., and Kelly-Quinn, M.: The impact of cattle access on ecological water quality in streams: Examples from agricultural catchments within Ireland, Sci. Total Environ., 547, 17–29, https://doi.org/10.1016/j.scitotenv.2015.12.120, 2016.
Coopersmith, E. J., Minsker, B., and Montagna, P.: Understanding and forecasting hypoxia using machine learning algorithms, J. Hydroinformatics, 13, 64, https://doi.org/10.2166/hydro.2010.015, 2010.
Coops, N. C., Waring, R. H., Beier, C., Roy-Jauvin, R., and Wang, T.: Modeling the occurrence of 15 coniferous tree species throughout the Pacific Northwest of North America using a hybrid approach of a generic process-based growth model and decision tree analysis, Appl. Veg. Sci., 14, 402–414, https://doi.org/10.1111/j.1654-109X.2011.01125.x, 2011.
Cozzi, S. and Giani, M.: River water and nutrient discharges in the Northern Adriatic Sea: Current importance and long term changes, Cont. Shelf Res., 31, 1881–1893, https://doi.org/10.1016/j.csr.2011.08.010, 2011.
Crowder, D. W., Demissie, M., and Markus, M.: The accuracy of sediment loads when log-transformation produces nonlinear sediment load-discharge relationships, J. Hydrol., 336, 250–268, https://doi.org/10.1016/j.jhydrol.2006.12.024, 2007.
Cutler, D. R., Edwards, T. C., Beard, K. H., Cutler, A., Hess, K. T., Gibson, J., and Lawler, J. J.: Random forests for classification in ecology, Ecology, 88, 2783–2792, https://doi.org/10.1890/07-0539.1, 2007.
Davies-Colley, R. J., Nagels, J. W., Smith, R. A., Young, R. G., and Phillips, C. J.: Water quality impact of a dairy cow herd crossing a stream, New Zeal. J. Mar. Freshw. Res., 38, 569–576, https://doi.org/10.1080/00288330.2004.9517262, 2004.
Denvil-Sommer, A., Gehlen, M., Vrac, M., and Mejia, C.: LSCE-FFNN-v1: a two-step neural network model for the reconstruction of surface ocean pCO2 over the global ocean, Geosci. Model Dev., 12, 2091–2105, https://doi.org/10.5194/gmd-12-2091-2019, 2019.
Domingues, R. B., Anselmo, T. P., Barbosa, A. B., Sommer, U., and Galvão, H. M.: Nutrient limitation of phytoplankton growth in the freshwater tidal zone of a turbid, Mediterranean estuary, Estuar. Coast. Shelf Sci., 91, 282–297, https://doi.org/10.1016/j.ecss.2010.10.033, 2011.
Erdal, H. I. and Karakurt, O.: Advancing monthly streamflow prediction accuracy of CART models using ensemble learning paradigms, J. Hydrol., 477, 119–128, https://doi.org/10.1016/j.jhydrol.2012.11.015, 2013.
Erlandsson, M., Cory, N., Fölster, J., Köhler, S., Laudon, H., Weyhenmeyer, G. A., and Bishop, K.: Increasing Dissolved Organic Carbon Redefines the Extent of Surface Water Acidification and Helps Resolve a Classic Controversy, Bioscience, 61, 614–618, https://doi.org/10.1525/bio.2011.61.8.7, 2011.
Filep, T. and Rékási, M.: Factors controlling dissolved organic carbon (DOC), dissolved organic nitrogen (DON) and DOC/DON ratio in arable soils based on a dataset from Hungary, Geoderma, 162, 312–318, https://doi.org/10.1016/j.geoderma.2011.03.002, 2011.
Forio, M. A. E., Landuyt, D., Bennetsen, E., Lock, K., Nguyen, T. H. T., Ambarita, M. N. D., Musonge, P. L. S., Boets, P., Everaert, G., Dominguez-Granda, L., and Goethals, P. L. M.: Bayesian belief network models to analyse and predict ecological water quality in rivers, Ecol. Modell., 312, 222–238, https://doi.org/10.1016/j.ecolmodel.2015.05.025, 2015.
Friedman, J.: Greedy Function Approximation: A Gradient Boosting Machine, Ann. Stat., 29, 1189–1232, https://doi.org/10.1214/009053606000000795, 2001.
Friedman, J. H.: Stochastic gradient boosting, Comput. Stat. Data Anal., 38, 367–378, https://doi.org/10.1016/S0167-9473(01)00065-2, 2002.
Fuka, D., Walter, T., Archibald, J., Tammo, S., and Easton, Z.: EcoHydRology: A Community Modeling Foundation for Eco-Hydrology, R package version 0.4.12.1, available at: https://cran.r-project.org/web/packages/EcoHydRology (last access: 9 September 2020), 2018.
Furey, P. R. and Gupta, V. K.: A physically based filter for separating base flow from streamflow time series, Water Resour. Res., 37, 2709–2722, https://doi.org/10.1029/2001WR000243, 2001.
Giblin, A. E., Weston, N. B., Banta, G. T., Tucker, J., and Hopkinson, C. S.: The Effects of Salinity on Nitrogen Losses from an Oligohaline Estuarine Sediment, Estuar. Coast., 33, 1054–1068, https://doi.org/10.1007/s12237-010-9280-7, 2010.
Górniak, A., Zieliński, P., Jekatierynczuk-Rudczyk, E., Grabowska, M. and Suchowolec, T.: The role of dissolved organic carbon in a shallow lowland reservoir ecosystem – A long-term study, Acta Hydrochim. Hydrobiol., 30, 179–189, https://doi.org/10.1002/aheh.200390001, 2002.
Green, C. T., Bekins, B. a, Kalkhoff, S. J., Hirsch, R. M., Liao, L., and Barnes, K. K.: Decadal surface water quality trends under variable climate, land use, and hydrogeochemical setting in Iowa, USA, Water Resour. Res., 50, 2425–2443, https://doi.org/10.1002/2013WR014829, 2014.
Greening, H., Janicki, A., Sherwood, E. T., Pribble, R., and Johansson, J. O. R.: Ecosystem responses to long-term nutrient management in an urban estuary: Tampa Bay, Florida, USA, Estuar. Coast. Shelf Sci., 151, A1–A16, https://doi.org/10.1016/j.ecss.2014.10.003, 2014.
Gunaratne, G. L., Vogwill, R. I. J., and Hipsey, M. R.: Effect of seasonal flushing on nutrient export characteristics of an urbanizing, remote, ungauged coastal catchment, Hydrol. Sci. J., 62, 800–817, https://doi.org/10.1080/02626667.2016.1264585, 2017.
Guo, D., Lintern, A., Webb, J. A., Ryu, D., Liu, S., Bende-Michl, U., Leahy, P., Wilson, P., and Western, A. W.: Key Factors Affecting Temporal Variability in Stream Water Quality, Water Resour. Res., 55, 112–129, https://doi.org/10.1029/2018WR023370, 2019.
Halliday, S. J., Wade, A. J., Skeffington, R. A., Neal, C., Reynolds, B., Rowland, P., Neal, M., and Norris, D.: An analysis of long-term trends, seasonality and short-term dynamics in water quality data from Plynlimon, Wales, Sci. Total Environ., 434, 186–200, https://doi.org/10.1016/j.scitotenv.2011.10.052, 2012.
Heathwaite, A. L.: Multiple stressors on water availability at global to catchment scales: Understanding human impact on nutrient cycles to protect water quality and water availability in the long term, Freshw. Biol., 55, 241–257, https://doi.org/10.1111/j.1365-2427.2009.02368.x, 2010.
Herndon, E. M., Dere, A. L., Sullivan, P. L., Norris, D., Reynolds, B., and Brantley, S. L.: Landscape heterogeneity drives contrasting concentration–discharge relationships in shale headwater catchments, Hydrol. Earth Syst. Sci., 19, 3333–3347, https://doi.org/10.5194/hess-19-3333-2015, 2015.
Hirsch, R. M. and De Cicco, L.: User guide to Exploration and Graphics for RivEr Trends (EGRET) and dataRetrieval: R packages for hydrologic data, Tech. Methods B, 4, 93, https://doi.org/10.3133/tm4A10, 2015.
Hirsch, R. M., Moyer, D. L., and Archfield, S. A.: Weighted regressions on time, discharge, and season (WRTDS), with an application to chesapeake bay river inputs, J. Am. Water Resour. Assoc., 46, 857–880, https://doi.org/10.1111/j.1752-1688.2010.00482.x, 2010.
Holguin-Gonzalez, J. E., Everaert, G., Boets, P., Galvis, A., and Goethals, P. L. M.: Development and application of an integrated ecological modelling framework to analyze the impact of wastewater discharges on the ecological water quality of rivers, Environ. Model. Softw., 48, 27–36, https://doi.org/10.1016/j.envsoft.2013.06.004, 2013.
Huang, P., Trayler, K., Wang, B., Saeed, A., Oldham, C., Busch, B., and Hipsey, M.: An integrated modelling system for water quality forecasting in an urban eutrophic estuary: The Swan-Canning Estuary virtual observatory, J. Mar. Syst., 199, 103218, https://doi.org/10.1016/j.jmarsys.2019.103218, 2019.
Hunter, J. M., Maier, H. R., Gibbs, M. S., Foale, E. R., Grosvenor, N. A., Harders, N. P., and Kikuchi-Miller, T. C.: Framework for developing hybrid process-driven, artificial neural network and regression models for salinity prediction in river systems, Hydrol. Earth Syst. Sci., 22, 2987–3006, https://doi.org/10.5194/hess-22-2987-2018, 2018.
Ishwaran, H. and Kogalur, U. B.: Consistency of random survival forests, Stat. Probab. Lett., 80, 1056–1064, https://doi.org/10.1016/j.spl.2010.02.020, 2010.
Ismail, R. and Mutanga, O.: A comparison of regression tree ensembles: Predicting Sirex noctilio induced water stress in Pinus patula forests of KwaZulu-Natal, South Africa, Int. J. Appl. Earth Obs. Geoinf., 12, S45–S51, https://doi.org/10.1016/j.jag.2009.09.004, 2010.
Jickells, T. D., Andrews, J. E., Parkes, D. J., Suratman, S., Aziz, A. A., and Hee, Y. Y.: Nutrient transport through estuaries: The importance of the estuarine geography, Estuar. Coast. Shelf Sci., 150, 215–229, https://doi.org/10.1016/j.ecss.2014.03.014, 2014.
Jordan, P. and Cassidy, R.: Technical Note: Assessing a 24/7 solution for monitoring water quality loads in small river catchments, Hydrol. Earth Syst. Sci., 15, 3093–3100, https://doi.org/10.5194/hess-15-3093-2011, 2011.
Kaiser, D., Unger, D., Qiu, G., Zhou, H., and Gan, H.: Natural and human influences on nutrient transport through a small subtropical Chinese estuary, Sci. Total Environ., 450–451, 92–107, https://doi.org/10.1016/j.scitotenv.2013.01.096, 2013.
Kelsey, P., Hall, J., Kitsios, A., Quinton, B., and Shakya, D.: Hydrological and nutrient modelling of the Swan-Canning coastal catchments, Water Science technical series, Department of Water, Western Australia., 2010.
Kelsey, P., Hall, J., Kretschmer, P., Quiton, B., and Shakya, D.: Hydrological and nutrient modelling of the Peel-Harvey catchment, Water Science Technical Series, Department of Water, Western Australia., 2011.
Lamsal, S., Grunwald, S., Bruland, G. L., Bliss, C. M., and Comerford, N. B.: Regional hybrid geospatial modeling of soil nitrate-nitrogen in the Santa Fe River Watershed, Geoderma, 135, 233–247, https://doi.org/10.1016/j.geoderma.2005.12.009, 2006.
Li, J.: Assessing spatial predictive models in the environmental sciences: Accuracy measures, data variation and variance explained, Environ. Model. Softw., 80, 1–8, https://doi.org/10.1016/j.envsoft.2016.02.004, 2016.
Li, M., Xu, K., Watanabe, M., and Chen, Z.: Long-term variations in dissolved silicate, nitrogen, and phosphorus flux from the Yangtze River into the East China Sea and impacts on estuarine ecosystem, Estuar. Coast. Shelf Sci., 71, 3–12, https://doi.org/10.1016/j.ecss.2006.08.013, 2007.
Li, M., Lee, Y. J., Testa, J. M., Li, Y., Ni, W., Kemp, W. M., and Di Toro, D. M.: What drives interannual variability of hypoxia in Chesapeake Bay: Climate forcing versus nutrient loading?, Geophys. Res. Lett., 43, 2127–2134, https://doi.org/10.1002/2015GL067334, 2016.
Li, R., Liu, S., Zhang, G., Ren, J., and Zhang, J.: Biogeochemistry of nutrients in an estuary affected by human activities: The Wanquan River estuary, eastern Hainan Island, China, Cont. Shelf Res., 57, 18–31, https://doi.org/10.1016/j.csr.2012.02.013, 2013.
Lintern, A., Webb, J. A., Ryu, D., Liu, S., Waters, D., Leahy, P., Bende-Michl, U., and Western, A. W.: What are the key catchment characteristics affecting spatial differences in riverine water quality?, Water Resour. Res., 54, 7252–7272, https://doi.org/10.1029/2017WR022172, 2018.
Liu, S. M., Li, L. W., Zhang, G. L., Liu, Z., Yu, Z., and Ren, J. L.: Impacts of human activities on nutrient transports in the Huanghe (Yellow River) estuary, J. Hydrol., 430–431, 103–110, https://doi.org/10.1016/j.jhydrol.2012.02.005, 2012.
Lloyd, C. E. M., Freer, J. E., Collins, A. L., Johnes, P. J., and Jones, J. I.: Methods for detecting change in hydrochemical time series in response to targeted pollutant mitigation in river catchments, J. Hydrol., 514, 297–312, https://doi.org/10.1016/j.jhydrol.2014.04.036, 2014.
Lyne, V. and Hollick, M.: Stochastic time-variable rainfall-runoff modelling, Institution of Engineers, Canberra, Australia, p. 89–93, 1979.
Maier, H. R., Kapelan, Z., Kasprzyk, J., Kollat, J., Matott, L. S., Cunha, M. C., Dandy, G. C., Gibbs, M. S., Keedwell, E., Marchi, A., Ostfeld, A., Savic, D., Solomatine, D. P., Vrugt, J. A., Zecchin, A. C., Minsker, B. S., Barbour, E. J., Kuczera, G., Pasha, F., Castelletti, A., Giuliani, M., and Reed, P. M.: Evolutionary algorithms and other metaheuristics in water resources: current status, research challenges and future directions, Environ. Model. Softw., 62, 271–299, 2014.
Makler-Pick, V., Gal, G., Gorfine, M., Hipsey, M. R., and Carmel, Y.: Sensitivity analysis for complex ecological models – A new approach, Environ. Model. Softw., 26, 124–134, https://doi.org/10.1016/j.envsoft.2010.06.010, 2011.
Martínez-Rojas, M., Marín, N., and Vila, M. A.: The role of information technologies to address data handling in construction project management, J. Comput. Civ. Eng., 30, 1–10, https://doi.org/10.1061/(ASCE)CP.1943-5487.0000538, 2015.
McBratney, A. B., Odeh, I. O. A., Bishop, T. F. A., Dunbar, M. S., and Shatar, T. M.: An overview of pedometric techniques for use in soil survey, Geoderma, 97, 293–327, https://doi.org/10.1016/S0016-7061(00)00043-4, 2000.
Mellander, P. E., Melland, A. R., Jordan, P., Wall, D. P., Murphy, P. N. C., and Shortle, G.: Quantifying nutrient transfer pathways in agricultural catchments using high temporal resolution data, Environ. Sci. Policy, 24, 44–57, https://doi.org/10.1016/j.envsci.2012.06.004, 2012.
Meshgi, A., Schmitter, P., Chui, T. F. M., and Babovic, V.: Development of a modular streamflow model to quantify runoff contributions from different land uses in tropical urban environments using Genetic Programming, J. Hydrol., 525, 711–723, https://doi.org/10.1016/j.jhydrol.2015.04.032, 2015.
Meybeck, M. and Moatar, F.: Daily variability of river concentrations and fluxes: Indicators based on the segmentation of the rating curve, Hydrol. Process., 26, 1188–1207, https://doi.org/10.1002/hyp.8211, 2012.
Moatar, F., Abbott, B. W., Minaudo, C., Curie, F., and Pinay, G.: Elemental properties, hydrology, and biology interact to shape concentration-discharge curves for carbon, nutrients, sediment, and major ions, Water Resour. Res., 53, 1270–1287, https://doi.org/10.1002/2016WR019635, 2016.
Nathan, R. J. and McMahon, T. A.: Evaluation of automated techniques for base flow and recession analyses, Water Resour. Res., 26, 1465–1473, https://doi.org/10.1029/WR026i007p01465, 1990.
Nice, H., Foulsham, G., Bree, M., and Sarah, E.: A baseline study of contaminants in the sediments of the Swan and Canning estuaries, Water Science technical series report no. 6, Department of Water, Western Australia, 2009.
Noori, N. and Kalin, L.: Coupling SWAT and ANN models for enhanced daily streamflow prediction, J. Hydrol., 533, 141–151, https://doi.org/10.1016/j.jhydrol.2015.11.050, 2016.
Petrone, K. C.: Catchment export of carbon, nitrogen, and phosphorus across an agro-urban land use gradient, Swan-Canning River system, southwestern Australia, J. Geophys. Res., 115, G01016, https://doi.org/10.1029/2009JG001051, 2010.
Petrone, K. C., Richards, J. S., and Grierson, P. F.: Bioavailability and composition of dissolved organic carbon and nitrogen in a near coastal catchment of south-western Australia, Biogeochemistry, 92, 27–40, 2009.
Povak, N. A., Hessburg, P. F., McDonnell, T. C., Reynolds, K. M., Sullivan, T. J., Salter, R. B., and Cosby, B. J.: Machine learning and linear regression models to predict catchment-level base cation weathering rates across the southern Appalachian Mountain region, USA, Water Resour. Res., 50, 2798–2814, https://doi.org/10.1002/2013WR014222, 2014.
Puissant, A., Rougier, S., and Stumpf, A.: Object-oriented mapping of urban trees using Random Forest classifiers, Int. J. Appl. Earth Obs. Geoinf., 26, 235–245, https://doi.org/10.1016/j.jag.2013.07.002, 2014.
Ruibal-Conti, A. L., Summers, R., Weaver, D., and Hipsey, M. R.: Hydro-climatological non-stationarity shifts patterns of nutrient delivery to an estuarine system, Hydrol. Earth Syst. Sci. Discuss., 10, 11035–11092, https://doi.org/10.5194/hessd-10-11035-2013, 2013.
Schnoor, J. L.: 4.1. Water quality and its sustainability introduction. in: Comprehensive Water Quality and Purification, edited by: Ahuja, S., Elsiever, Waltham, pp. 1–40, 2014.
Seitzinger, S. P., Sanders, R. W., and Styles, R.: Bioavailability of DON from natural and anthropogenic sources to estuarine plankton, Limnol. Oceanogr., 47, 353–366, https://doi.org/10.4319/lo.2002.47.2.0353, 2002.
Singh, K. P., Gupta, S., and Mohan, D.: Evaluating influences of seasonal variations and anthropogenic activities on alluvial groundwater hydrochemistry using ensemble learning approaches, J. Hydrol., 511, 254–266, 2014.
Staehr, P. A., Testa, J., and Carstensen, J.: Decadal Changes in Water Quality and Net Productivity of a Shallow Danish Estuary Following Significant Nutrient Reductions, Estuar. Coast., 40, 63–79, https://doi.org/10.1007/s12237-016-0117-x, 2017.
Stallard, R. F. and Murphy, S. F.: A Unified Assessment of Hydrologic and Biogeochemical Responses in Research Watersheds in Eastern Puerto Rico Using Runoff-Concentration Relations, Aquat. Geochemistry, 20, 115–139, https://doi.org/10.1007/s10498-013-9216-5, 2014.
Swan River Trust: Swan Canning Water Quality Improvement., 2009.
Szilagyi, J. and Parlange, M. B.: Baseflow separation based on analytical solutions of the Boussinesq equation, J. Hydrol., 204, 251–260, https://doi.org/10.1016/S0022-1694(97)00132-7, 1998.
Tao, Y., Wei, M., Ongley, E., Li, Z., and Jingsheng, C.: Long-term variations and causal factors in nitrogen and phosphorus transport in the Yellow River, China, Estuar. Coast. Shelf Sci., 86, 345–351, https://doi.org/10.1016/j.ecss.2009.05.014, 2010.
Tesoriero, A. J., Duff, J. H., Wolock, D. M., Spahr, N. E., and Almendinger, J. E.: Identifying Pathways and Processes Affecting Nitrate and Orthophosphate Inputs to Streams in Agricultural Watersheds, J. Environ. Qual., 38, 1892, https://doi.org/10.2134/jeq2008.0484, 2009.
Testa, J. M., Clark, J. B., Dennison, W. C., Donovan, E. C., Fisher, A. W., Ni, W., Parker, M., Scavia, D., Spitzer, S. E., Waldrop, A. M., Vargas, V. M. D., and Ziegler, G.: Ecological Forecasting and the Science of Hypoxia in Chesapeake Bay, Bioscience, 67, 614–626, https://doi.org/10.1093/biosci/bix048, 2017.
Wang, B.: benyawang-uwa/daily-nutrient-prediction: first release of daily nutrient prediction model (Version v1.0.0), Zenodo, https://doi.org/10.5281/zenodo.3739611, 2020.
Wang, B., Hipsey, M. R., Ahmed, S., and Oldham, C.: The Impact of Landscape Characteristics on Groundwater Dissolved Organic Nitrogen: Insights From Machine Learning Methods and Sensitivity Analysis, Water Resour. Res., 54, 4785–4804, https://doi.org/10.1029/2017WR021749, 2018.
Yang, P., Yang, Y. H., Zhou, B. B., and Zomaya, A. Y.: A Review of Ensemble Methods in Bioinformatics, Curr. Bioinf., 5, 296–308, https://doi.org/10.2174/157489310794072508, 2010.
Zhang, Q., Harman, C. J., and Ball, W. P.: An improved method for interpretation of riverine concentration-discharge relationships indicates long-term shifts in reservoir sediment trapping, Geophys. Res. Lett., 43, 10215–10224, https://doi.org/10.1002/2016GL069945, 2016a.
Zhang, Q., Ball, W. P., and Moyer, D. L.: Decadal-scale export of nitrogen, phosphorus, and sediment from the Susquehanna River basin, USA: Analysis and synthesis of temporal and spatial patterns, Sci. Total Environ., 563–564, 1016–1029, https://doi.org/10.1016/j.scitotenv.2016.03.104, 2016b.
Zhang, Q., Hirsch, R. M., and Ball, W. P.: Long-Term Changes in Sediment and Nutrient Delivery from Conowingo Dam to Chesapeake Bay: Effects of Reservoir Sedimentation, Environ. Sci. Technol., 50, 1877–1886, https://doi.org/10.1021/acs.est.5b04073, 2016c.